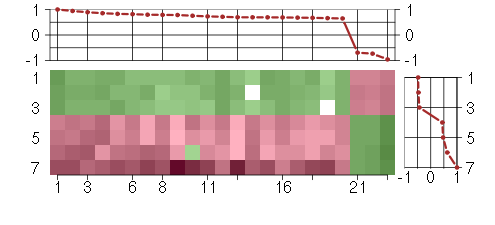

Under-expression is coded with green,
over-expression with red color.



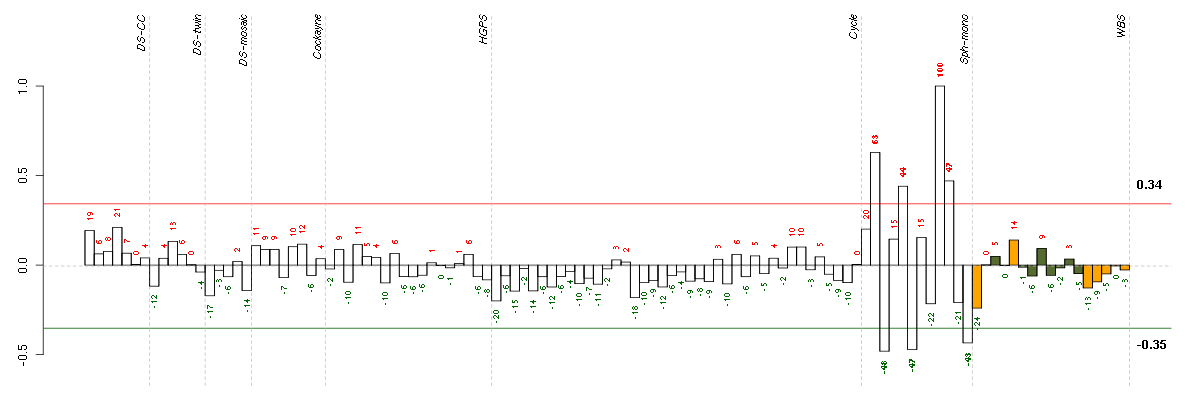
ACSL5acyl-CoA synthetase long-chain family member 5 (218322_s_at), score: 0.66 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.94 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: 0.73 BMP7bone morphogenetic protein 7 (209590_at), score: 0.69 C14orf169chromosome 14 open reading frame 169 (219526_at), score: -0.69 C20orf39chromosome 20 open reading frame 39 (219310_at), score: 0.79 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: 0.82 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: 0.71 EGLN3egl nine homolog 3 (C. elegans) (219232_s_at), score: 0.67 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: 0.8 GABRA2gamma-aminobutyric acid (GABA) A receptor, alpha 2 (207014_at), score: 0.78 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: -0.96 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: 0.69 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: 0.85 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 1 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: 0.83 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: 0.89 MYO1Bmyosin IB (212364_at), score: -0.73 NPTX1neuronal pentraxin I (204684_at), score: 0.65 RARBretinoic acid receptor, beta (205080_at), score: 0.69 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: 0.69 SULT1A1sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (203615_x_at), score: 0.68 VAV3vav 3 guanine nucleotide exchange factor (218807_at), score: 0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |