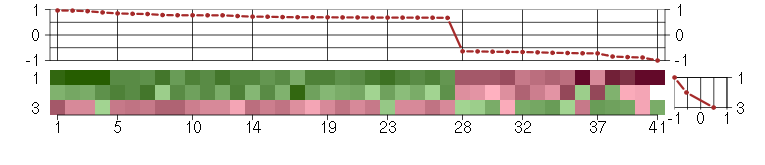

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
small GTPase regulator activity
Modulates the rate of GTP hydrolysis by a small monomeric GTPase.
GTPase activator activity
Increases the activity of a GTPase, an enzyme that catalyzes the hydrolysis of GTP.
Ras GTPase activator activity
Increases the rate of GTP hydrolysis by a GTPase of the Ras superfamily.
enzyme activator activity
Increases the activity of an enzyme.
enzyme regulator activity
Modulates the activity of an enzyme.
GTPase regulator activity
Modulates the rate of GTP hydrolysis by a GTPase.
Rap GTPase activator activity
Increases the rate of GTP hydrolysis by a GTPase of the Rap family.
all
This term is the most general term possible
GTPase activator activity
Increases the activity of a GTPase, an enzyme that catalyzes the hydrolysis of GTP.
Ras GTPase activator activity
Increases the rate of GTP hydrolysis by a GTPase of the Ras superfamily.
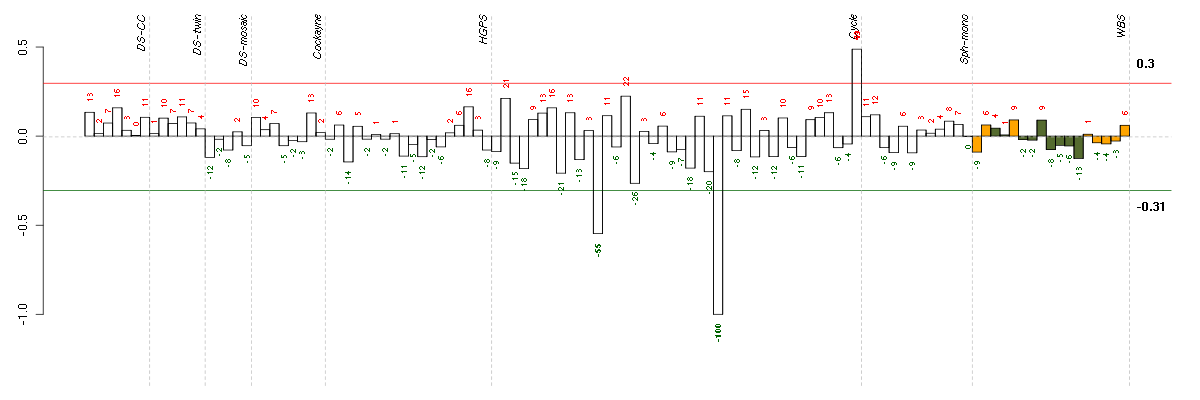
AGMATagmatine ureohydrolase (agmatinase) (219792_at), score: -0.84 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: 0.85 B3GALT4UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (210205_at), score: 0.93 C2orf37chromosome 2 open reading frame 37 (220172_at), score: -0.72 C6orf35chromosome 6 open reading frame 35 (218453_s_at), score: -0.64 DOK4docking protein 4 (209691_s_at), score: -0.7 DYMdymeclin (220774_at), score: 0.72 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: 0.68 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: 0.77 FBXO17F-box protein 17 (220233_at), score: -0.68 FLCNfolliculin (215645_at), score: 0.72 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: 0.77 HIST1H2ALhistone cluster 1, H2al (214554_at), score: -0.88 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.68 JAG2jagged 2 (32137_at), score: 0.74 KRT86keratin 86 (215189_at), score: 0.78 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.7 MAGI1membrane associated guanylate kinase, WW and PDZ domain containing 1 (206144_at), score: -1 MGAT4Cmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) (207447_s_at), score: -0.72 NOS1nitric oxide synthase 1 (neuronal) (207309_at), score: 0.7 OLIG2oligodendrocyte lineage transcription factor 2 (213825_at), score: -0.65 OR1E2olfactory receptor, family 1, subfamily E, member 2 (208587_s_at), score: 0.82 PLA2G7phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) (206214_at), score: 0.96 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.7 PPP1R14Dprotein phosphatase 1, regulatory (inhibitor) subunit 14D (220082_at), score: 0.68 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -0.71 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: 0.77 SAA4serum amyloid A4, constitutive (207096_at), score: 0.67 SARM1sterile alpha and TIR motif containing 1 (213259_s_at), score: 0.68 SHPKsedoheptulokinase (219713_at), score: 0.89 SIPA1signal-induced proliferation-associated 1 (204164_at), score: -0.67 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: 0.69 SMA4glucuronidase, beta pseudogene (215599_at), score: -0.87 SP140SP140 nuclear body protein (207777_s_at), score: -0.64 SPAG8sperm associated antigen 8 (206816_s_at), score: 0.83 TBX10T-box 10 (207689_at), score: 0.69 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: 0.77 TNFRSF8tumor necrosis factor receptor superfamily, member 8 (206729_at), score: 0.96 WDR78WD repeat domain 78 (220769_s_at), score: 0.68 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: 0.69 ZNF510zinc finger protein 510 (206053_at), score: -0.66
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |