

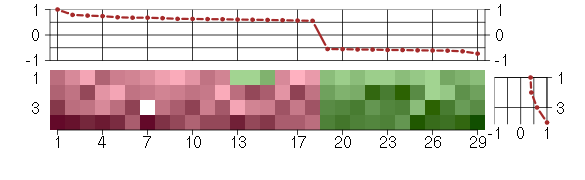

Under-expression is coded with green,
over-expression with red color.



ARHGEF11Rho guanine nucleotide exchange factor (GEF) 11 (202914_s_at), score: 0.62 C1orf159chromosome 1 open reading frame 159 (219337_at), score: 0.6 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: 1 DNM3dynamin 3 (209839_at), score: 0.59 DUSP6dual specificity phosphatase 6 (208892_s_at), score: 0.58 EDN1endothelin 1 (218995_s_at), score: 0.76 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: 0.74 EGR3early growth response 3 (206115_at), score: 0.69 EMCNendomucin (219436_s_at), score: -0.55 IER3immediate early response 3 (201631_s_at), score: 0.63 JUNBjun B proto-oncogene (201473_at), score: 0.79 KIAA0907KIAA0907 (202220_at), score: -0.62 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: 0.62 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.56 OCLMoculomedin (208274_at), score: -0.6 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.63 RABL3RAB, member of RAS oncogene family-like 3 (213970_at), score: -0.61 RARAretinoic acid receptor, alpha (203749_s_at), score: 0.68 RPL10Lribosomal protein L10-like (217559_at), score: -0.59 RSBN1round spermatid basic protein 1 (213694_at), score: -0.73 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: 0.61 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (218985_at), score: 0.66 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: -0.56 WDR5BWD repeat domain 5B (219538_at), score: -0.57 WDR76WD repeat domain 76 (205519_at), score: 0.57 ZC3H12Azinc finger CCCH-type containing 12A (218810_at), score: 0.68 ZNF280Dzinc finger protein 280D (221213_s_at), score: -0.59 ZNF337zinc finger protein 337 (37860_at), score: -0.64 ZNF75Dzinc finger protein 75D (213659_at), score: -0.57
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| 9837_CNTL.CEL | 12 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
