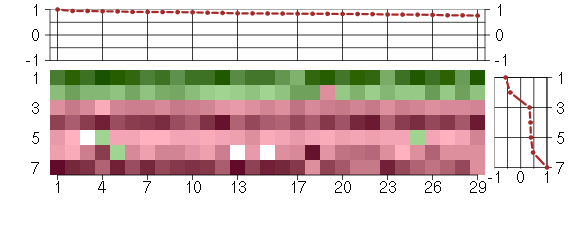

Under-expression is coded with green,
over-expression with red color.



ACSL6acyl-CoA synthetase long-chain family member 6 (211207_s_at), score: 0.83 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: 0.89 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: 0.91 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.79 CDH19cadherin 19, type 2 (206898_at), score: 1 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: 0.76 CHN2chimerin (chimaerin) 2 (207486_x_at), score: 0.85 CYCSP52cytochrome c, somatic pseudogene 52 (217206_at), score: 0.8 DHRS9dehydrogenase/reductase (SDR family) member 9 (219799_s_at), score: 0.82 DTNBdystrobrevin, beta (215295_at), score: 0.9 ICA1islet cell autoantigen 1, 69kDa (211740_at), score: 0.9 KCNIP1Kv channel interacting protein 1 (221307_at), score: 0.84 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.82 LHX3LIM homeobox 3 (221670_s_at), score: 0.82 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: 0.85 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: 0.9 MYO1Amyosin IA (211916_s_at), score: 0.93 MYO5Cmyosin VC (218966_at), score: 0.94 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.8 PCDH11Yprotocadherin 11 Y-linked (211227_s_at), score: 0.84 PDE3Aphosphodiesterase 3A, cGMP-inhibited (206388_at), score: 0.92 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: 0.77 PRLprolactin (205445_at), score: 0.84 RP11-35N6.1plasticity related gene 3 (219732_at), score: 0.87 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: 0.8 TAAR2trace amine associated receptor 2 (221394_at), score: 0.94 TLR1toll-like receptor 1 (210176_at), score: 0.87 TSPY1testis specific protein, Y-linked 1 (216374_at), score: 0.77 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.83
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |