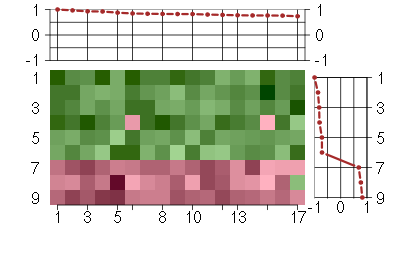

Under-expression is coded with green,
over-expression with red color.



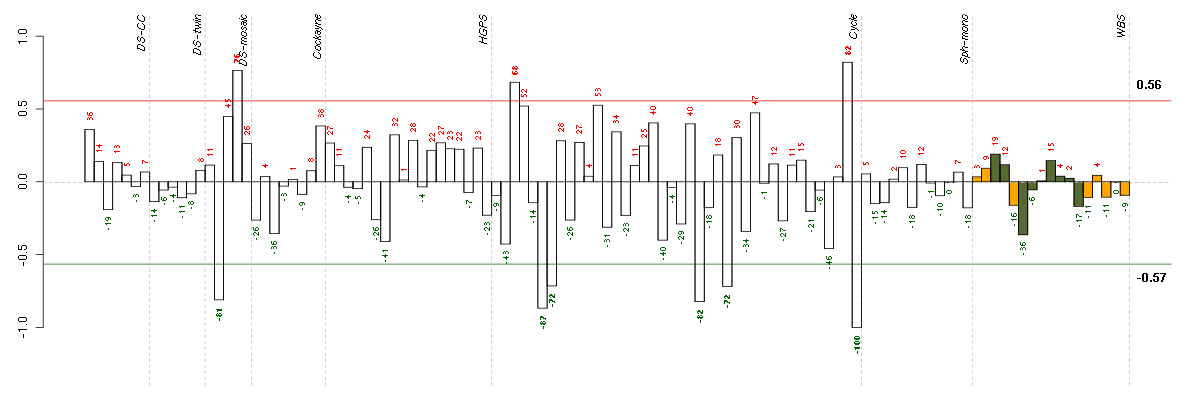
ADCY3adenylate cyclase 3 (209321_s_at), score: 0.73 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.77 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.76 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.83 DSC3desmocollin 3 (206032_at), score: 0.88 FKRPfukutin related protein (219853_at), score: 0.83 FOXK2forkhead box K2 (203064_s_at), score: 0.84 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.77 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.8 NDST1N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 (202608_s_at), score: 0.79 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.82 PXNpaxillin (211823_s_at), score: 0.93 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 1 SOCS1suppressor of cytokine signaling 1 (210001_s_at), score: 0.82 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.97 TSKUtsukushin (218245_at), score: 0.77 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.92
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |