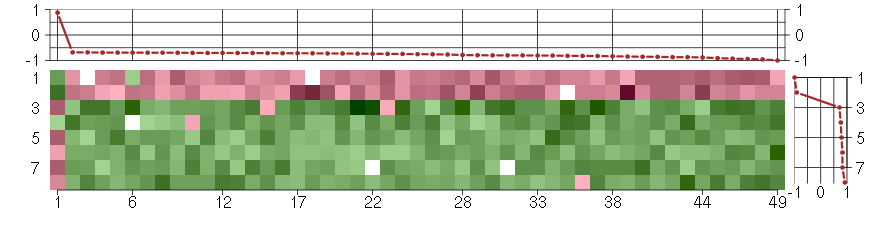

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

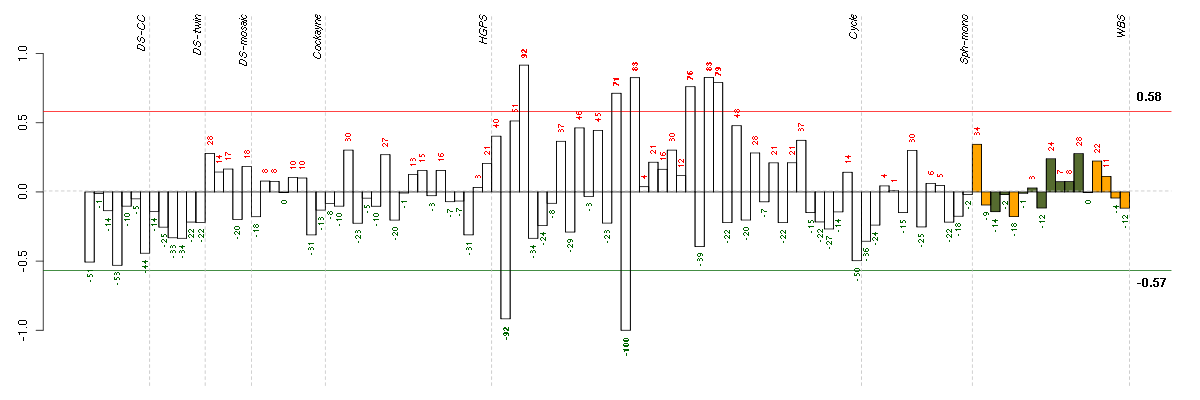
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.85 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: -0.72 ADORA1adenosine A1 receptor (216220_s_at), score: -0.69 APPamyloid beta (A4) precursor protein (214953_s_at), score: -0.74 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: -0.72 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: -0.72 C4Acomplement component 4A (Rodgers blood group) (214428_x_at), score: -0.7 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: -0.75 CYTH4cytohesin 4 (219183_s_at), score: -0.72 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.94 EDAectodysplasin A (211130_x_at), score: -0.85 FBRSfibrosin (218255_s_at), score: -0.82 FBXO2F-box protein 2 (219305_x_at), score: -0.75 FGL1fibrinogen-like 1 (205305_at), score: -0.71 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.88 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: -0.83 GPR144G protein-coupled receptor 144 (216289_at), score: -1 GRM1glutamate receptor, metabotropic 1 (210939_s_at), score: -0.69 HAB1B1 for mucin (215778_x_at), score: -0.91 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.68 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.69 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: -0.82 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (217039_x_at), score: -0.74 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: -0.96 KIF26Bkinesin family member 26B (220002_at), score: -0.69 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: -0.8 LOC149478hypothetical protein LOC149478 (215462_at), score: -0.71 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.8 PGCprogastricsin (pepsinogen C) (205261_at), score: -0.72 PKLRpyruvate kinase, liver and RBC (222078_at), score: -0.93 PMS2L2postmeiotic segregation increased 2-like 2 pseudogene (215410_at), score: -0.81 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.87 RAB40BRAB40B, member RAS oncogene family (217597_x_at), score: -0.73 RBM38RNA binding motif protein 38 (212430_at), score: -0.77 RNF121ring finger protein 121 (219021_at), score: -0.7 SAA4serum amyloid A4, constitutive (207096_at), score: -0.69 SEPT5septin 5 (209767_s_at), score: -0.82 SH2D3ASH2 domain containing 3A (222169_x_at), score: -0.8 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.79 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: 0.88 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.71 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: -0.86 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: -0.87 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: -0.7 TMPRSS6transmembrane protease, serine 6 (214955_at), score: -0.79 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.76 TNFRSF8tumor necrosis factor receptor superfamily, member 8 (206729_at), score: -0.81 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: -0.7 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.68
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |