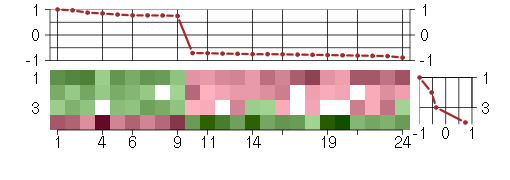

Under-expression is coded with green,
over-expression with red color.



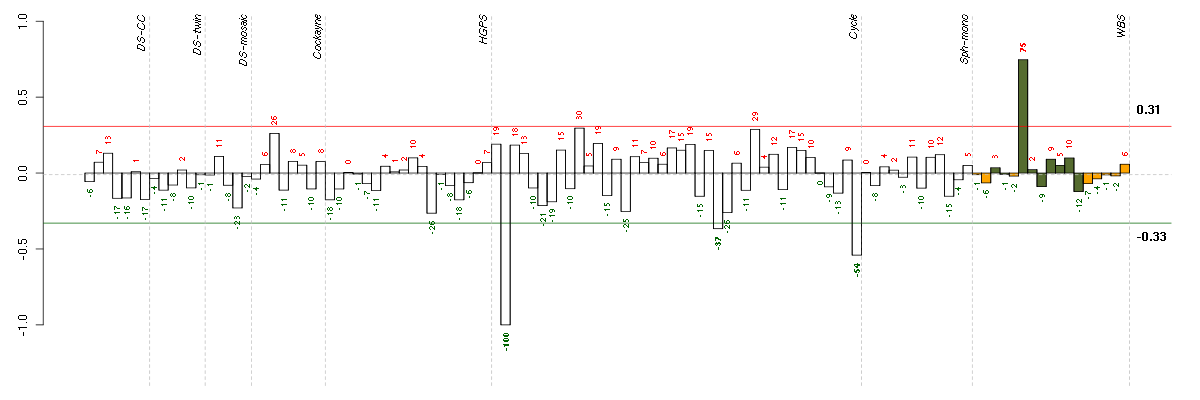
C11orf30chromosome 11 open reading frame 30 (219012_s_at), score: 0.77 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: -0.89 FAM115Afamily with sequence similarity 115, member A (212981_s_at), score: -0.79 FUT4fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) (209892_at), score: 0.77 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (209631_s_at), score: 0.75 HIRAHIR histone cell cycle regulation defective homolog A (S. cerevisiae) (217427_s_at), score: -0.8 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.81 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.82 KCTD17potassium channel tetramerisation domain containing 17 (205561_at), score: -0.77 KRT8P12keratin 8 pseudogene 12 (222060_at), score: -0.78 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.71 LOC149501similar to keratin 8 (216821_at), score: -0.83 LOC222070hypothetical protein LOC222070 (221949_at), score: 0.77 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: 1 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.76 PDIA3protein disulfide isomerase family A, member 3 (208612_at), score: -0.75 PHF2PHD finger protein 2 (212726_at), score: 0.88 PTP4A1protein tyrosine phosphatase type IVA, member 1 (200730_s_at), score: -0.71 RBM38RNA binding motif protein 38 (212430_at), score: -0.75 REG3Aregenerating islet-derived 3 alpha (205815_at), score: -0.74 RNF146ring finger protein 146 (221430_s_at), score: -0.73 RUNDC3BRUN domain containing 3B (215321_at), score: 0.85 TTC30Atetratricopeptide repeat domain 30A (213679_at), score: 0.8 ZNF3zinc finger protein 3 (212684_at), score: 0.97
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 5042_CNTL.CEL | 6 | 8 | WBS | hgu133plus2 | none | WBS 1 |