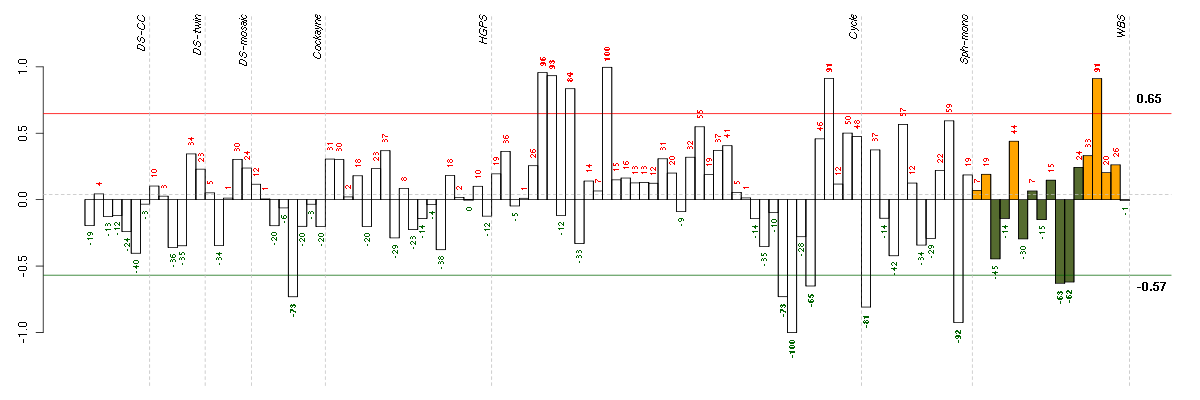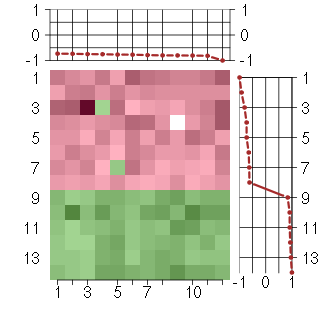

Under-expression is coded with green,
over-expression with red color.



CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213348_at), score: -1 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.8 CTDSP2CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 (203445_s_at), score: -0.73 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: -0.81 LOC100132540similar to LOC339047 protein (214870_x_at), score: -0.75 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.8 LOC399491LOC399491 protein (214035_x_at), score: -0.82 PELI2pellino homolog 2 (Drosophila) (219132_at), score: -0.75 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.77 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: -0.79 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.77 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: -0.74
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 9118_CNTL.CEL | 11 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |