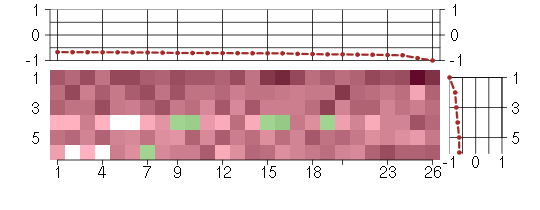

Under-expression is coded with green,
over-expression with red color.



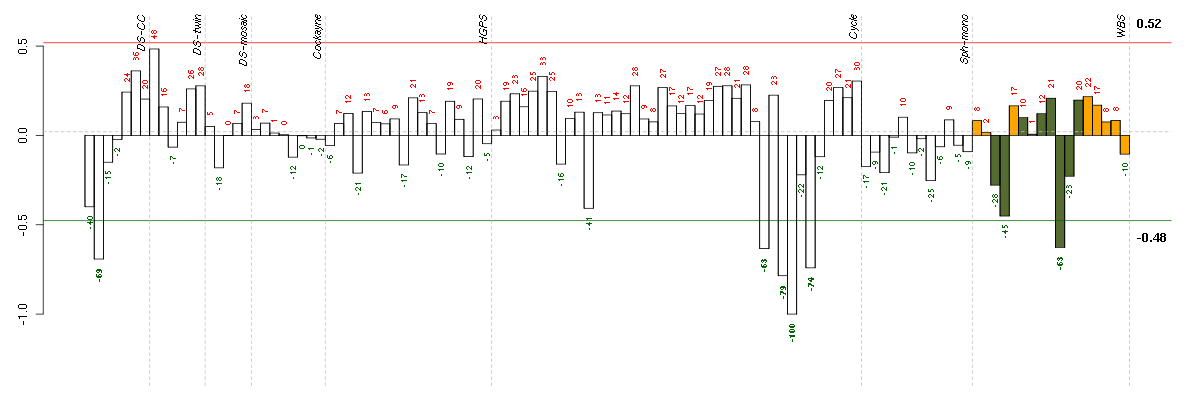
ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: -0.91 ANGPTL2angiopoietin-like 2 (213001_at), score: -0.68 ANK2ankyrin 2, neuronal (202920_at), score: -0.75 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: -0.77 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: -0.72 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: -0.68 DAPK1death-associated protein kinase 1 (203139_at), score: -0.77 ECM2extracellular matrix protein 2, female organ and adipocyte specific (206101_at), score: -0.69 ENOX1ecto-NOX disulfide-thiol exchanger 1 (219501_at), score: -0.76 EPHB6EPH receptor B6 (204718_at), score: -0.71 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: -0.67 FRYfurry homolog (Drosophila) (204072_s_at), score: -0.72 IFIH1interferon induced with helicase C domain 1 (219209_at), score: -0.71 KCNB1potassium voltage-gated channel, Shab-related subfamily, member 1 (211006_s_at), score: -0.76 KCND3potassium voltage-gated channel, Shal-related subfamily, member 3 (213832_at), score: -0.71 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: -0.69 LZTFL1leucine zipper transcription factor-like 1 (218437_s_at), score: -0.67 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: -0.79 MX2myxovirus (influenza virus) resistance 2 (mouse) (204994_at), score: -0.72 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: -0.68 PPLperiplakin (203407_at), score: -1 PRKG1protein kinase, cGMP-dependent, type I (207119_at), score: -0.68 SQLEsqualene epoxidase (213562_s_at), score: -0.71 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.79 TNXAtenascin XA pseudogene (213451_x_at), score: -0.74 TNXBtenascin XB (216333_x_at), score: -0.71
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |