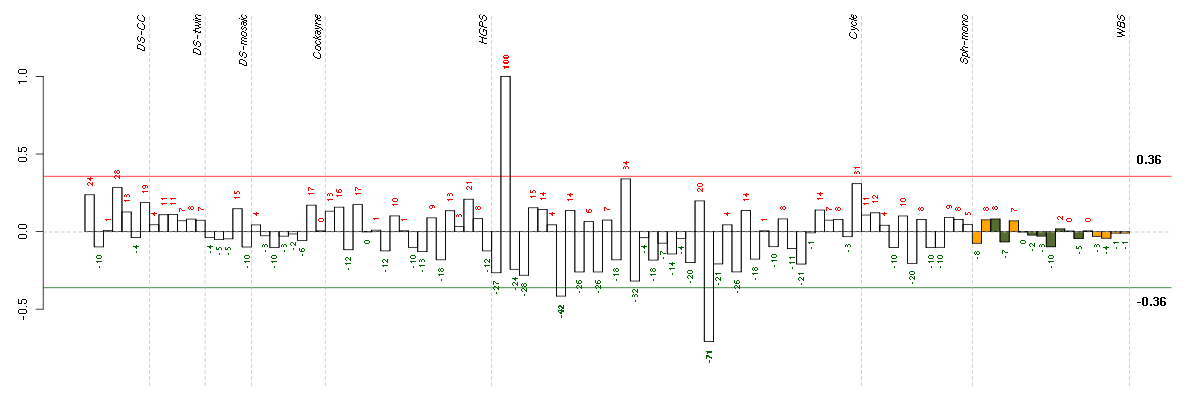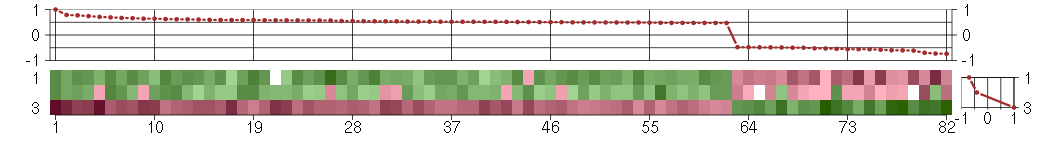

Under-expression is coded with green,
over-expression with red color.

transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells.
cation transport
The directed movement of cations, atoms or small molecules with a net positive charge, into, out of, within or between cells.
metal ion transport
The directed movement of metal ions, any metal ion with an electric charge, into, out of, within or between cells.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
all
This term is the most general term possible
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.


ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 1 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.64 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: 0.57 APPamyloid beta (A4) precursor protein (214953_s_at), score: 0.57 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.79 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: 0.47 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.57 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: -0.61 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: 0.59 CACNA1Gcalcium channel, voltage-dependent, T type, alpha 1G subunit (211315_s_at), score: 0.51 CATSPER2cation channel, sperm associated 2 (217588_at), score: 0.62 CC2D1Acoiled-coil and C2 domain containing 1A (58994_at), score: 0.49 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.52 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.51 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: 0.47 CYorf15Bchromosome Y open reading frame 15B (214131_at), score: 0.66 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.54 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: -0.5 EPN1epsin 1 (221141_x_at), score: 0.51 EPOerythropoietin (217254_s_at), score: 0.49 EPS8L1EPS8-like 1 (218778_x_at), score: 0.61 ETNK2ethanolamine kinase 2 (219268_at), score: 0.58 EXPH5exophilin 5 (214734_at), score: -0.48 FGF13fibroblast growth factor 13 (205110_s_at), score: -0.49 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.74 FXYD2FXYD domain containing ion transport regulator 2 (207434_s_at), score: 0.55 GABRR2gamma-aminobutyric acid (GABA) receptor, rho 2 (208217_at), score: 0.52 GASTgastrin (208138_at), score: 0.55 GATMglycine amidinotransferase (L-arginine:glycine amidinotransferase) (203178_at), score: -0.7 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.53 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: 0.5 HIST1H4Ghistone cluster 1, H4g (208551_at), score: 0.58 HOXA4homeobox A4 (206289_at), score: 0.47 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: -0.53 HSPA6heat shock 70kDa protein 6 (HSP70B') (117_at), score: 0.54 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: 0.47 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.64 KCTD17potassium channel tetramerisation domain containing 17 (205561_at), score: 0.57 KIF16Bkinesin family member 16B (219570_at), score: -0.6 KIF26Bkinesin family member 26B (220002_at), score: 0.52 KRT8P12keratin 8 pseudogene 12 (222060_at), score: 0.71 LOC100128919similar to HSPC157 (219865_at), score: -0.74 LOC100133748similar to GTF2IRD2 protein (215569_at), score: 0.62 LSRlipolysis stimulated lipoprotein receptor (208190_s_at), score: -0.5 LTBP4latent transforming growth factor beta binding protein 4 (210628_x_at), score: 0.5 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.5 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (213178_s_at), score: 0.49 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: -0.56 MGC13053hypothetical MGC13053 (211775_x_at), score: -0.58 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.61 NPEPL1aminopeptidase-like 1 (89476_r_at), score: 0.49 NUPL2nucleoporin like 2 (204003_s_at), score: 0.52 OPRL1opiate receptor-like 1 (206564_at), score: 0.52 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: 0.47 PAX4paired box 4 (207867_at), score: -0.56 PCOTHprostate collagen triple helix (222277_at), score: -0.52 PGK2phosphoglycerate kinase 2 (217009_at), score: 0.51 PHF2PHD finger protein 2 (212726_at), score: -0.6 PMS2L2postmeiotic segregation increased 2-like 2 pseudogene (215410_at), score: 0.59 PNMAL1PNMA-like 1 (218824_at), score: -0.48 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.57 RBAKRB-associated KRAB zinc finger (219214_s_at), score: 0.49 RBM12BRNA binding motif protein 12B (51228_at), score: -0.56 RBM38RNA binding motif protein 38 (212430_at), score: 0.59 RNF121ring finger protein 121 (219021_at), score: 0.54 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: 0.48 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: 0.67 SCLYselenocysteine lyase (221575_at), score: -0.73 SIX6SIX homeobox 6 (207250_at), score: 0.69 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: -0.48 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: -0.54 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.59 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.47 STXBP2syntaxin binding protein 2 (209367_at), score: 0.48 SYT5synaptotagmin V (206161_s_at), score: 0.59 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.54 TMPRSS6transmembrane protease, serine 6 (214955_at), score: 0.77 TRMUtRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (213634_s_at), score: 0.52 TRPV4transient receptor potential cation channel, subfamily V, member 4 (219516_at), score: 0.49 WFDC8WAP four-disulfide core domain 8 (215276_at), score: 0.52 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.53 ZNF192zinc finger protein 192 (206579_at), score: -0.49
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |