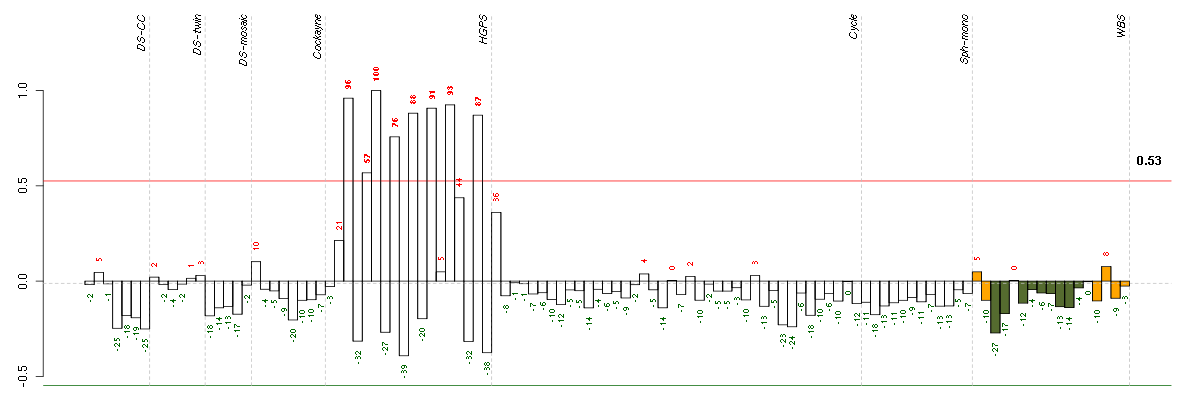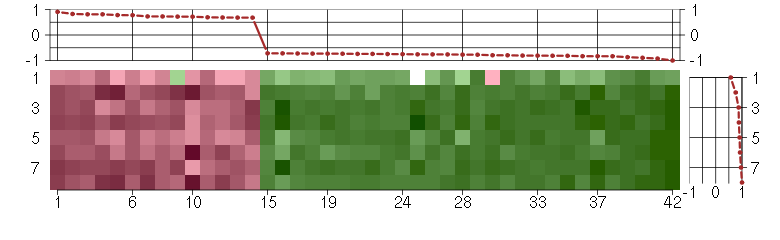

Under-expression is coded with green,
over-expression with red color.



ANKRD1ankyrin repeat domain 1 (cardiac muscle) (206029_at), score: -0.92 AUTS2autism susceptibility candidate 2 (212599_at), score: 0.73 BST1bone marrow stromal cell antigen 1 (205715_at), score: -0.72 C1QTNF3C1q and tumor necrosis factor related protein 3 (220988_s_at), score: 0.72 CAMK2N1calcium/calmodulin-dependent protein kinase II inhibitor 1 (218309_at), score: 0.91 CDH4cadherin 4, type 1, R-cadherin (retinal) (206866_at), score: 0.68 EPHA5EPH receptor A5 (215664_s_at), score: -0.89 F2Rcoagulation factor II (thrombin) receptor (203989_x_at), score: -0.83 GPR116G protein-coupled receptor 116 (212950_at), score: -0.81 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: 0.78 IFI27interferon, alpha-inducible protein 27 (202411_at), score: -0.73 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.69 JAG1jagged 1 (Alagille syndrome) (216268_s_at), score: -0.81 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: 0.73 KRT18keratin 18 (201596_x_at), score: -0.77 LIPGlipase, endothelial (219181_at), score: -0.81 LMO2LIM domain only 2 (rhombotin-like 1) (204249_s_at), score: -0.8 LPHN2latrophilin 2 (206953_s_at), score: -0.83 LPXNleupaxin (216250_s_at), score: 0.68 LRRC15leucine rich repeat containing 15 (213909_at), score: 0.81 LRRC17leucine rich repeat containing 17 (205381_at), score: -0.73 MBPmyelin basic protein (210136_at), score: -0.76 MEOX2mesenchyme homeobox 2 (206201_s_at), score: -1 MESTmesoderm specific transcript homolog (mouse) (202016_at), score: -0.82 MGPmatrix Gla protein (202291_s_at), score: -0.83 NDUFA4L2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 (218484_at), score: -0.79 NET1neuroepithelial cell transforming 1 (201830_s_at), score: 0.69 NKX3-1NK3 homeobox 1 (209706_at), score: 0.78 PDE10Aphosphodiesterase 10A (205501_at), score: -0.75 PLA2G16phospholipase A2, group XVI (209581_at), score: -0.76 PNMA2paraneoplastic antigen MA2 (209598_at), score: -0.74 POSTNperiostin, osteoblast specific factor (210809_s_at), score: -0.72 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: -0.77 RELNreelin (205923_at), score: -0.75 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: 0.81 SRGNserglycin (201859_at), score: -0.76 TINAGL1tubulointerstitial nephritis antigen-like 1 (219058_x_at), score: -0.72 TMEM158transmembrane protein 158 (213338_at), score: 0.83 TNXAtenascin XA pseudogene (213451_x_at), score: -0.74 TNXBtenascin XB (216333_x_at), score: -0.73 ZNF423zinc finger protein 423 (214761_at), score: -0.87 ZNF536zinc finger protein 536 (206403_at), score: 0.72
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690223.cel | 3 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |