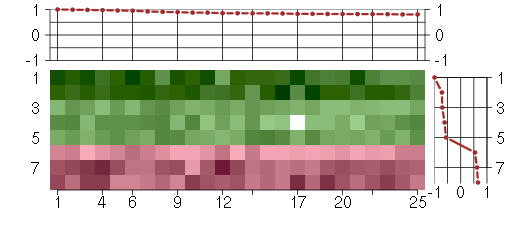

Under-expression is coded with green,
over-expression with red color.



ATP6V1AATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (201971_s_at), score: 0.82 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: 0.95 CDV3CDV3 homolog (mouse) (213548_s_at), score: 0.84 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.85 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: 1 CPDcarboxypeptidase D (201942_s_at), score: 0.88 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: 0.98 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.97 FN1fibronectin 1 (214701_s_at), score: 0.92 GNSglucosamine (N-acetyl)-6-sulfatase (203676_at), score: 0.82 GPR137G protein-coupled receptor 137 (43934_at), score: 0.85 GTF2Igeneral transcription factor II, i (210892_s_at), score: 0.85 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: 0.82 IL6STinterleukin 6 signal transducer (gp130, oncostatin M receptor) (204864_s_at), score: 0.82 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: 0.82 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.96 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.99 PHTF2putative homeodomain transcription factor 2 (217097_s_at), score: 0.83 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: 0.91 PXNpaxillin (211823_s_at), score: 0.81 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: 0.85 RXRBretinoid X receptor, beta (215099_s_at), score: 0.81 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: 0.83 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.88 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: 0.89
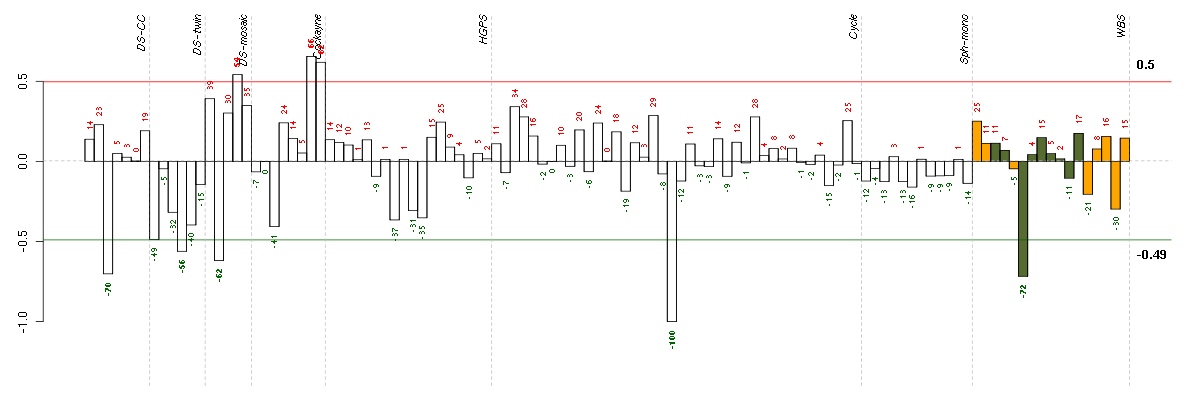
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 5042_CNTL.CEL | 6 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |