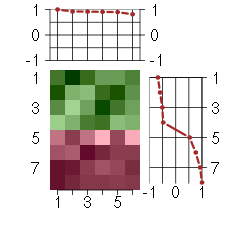

Under-expression is coded with green,
over-expression with red color.



CDKN2AIPCDKN2A interacting protein (218929_at), score: 0.92 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: 0.81 NPATnuclear protein, ataxia-telangiectasia locus (209798_at), score: 0.9 PHF15PHD finger protein 15 (212660_at), score: 0.91 RGNEFRho-guanine nucleotide exchange factor (219610_at), score: 1 WHAMML1WAS protein homolog associated with actin, golgi membranes and microtubules-like 1 (213908_at), score: 0.92
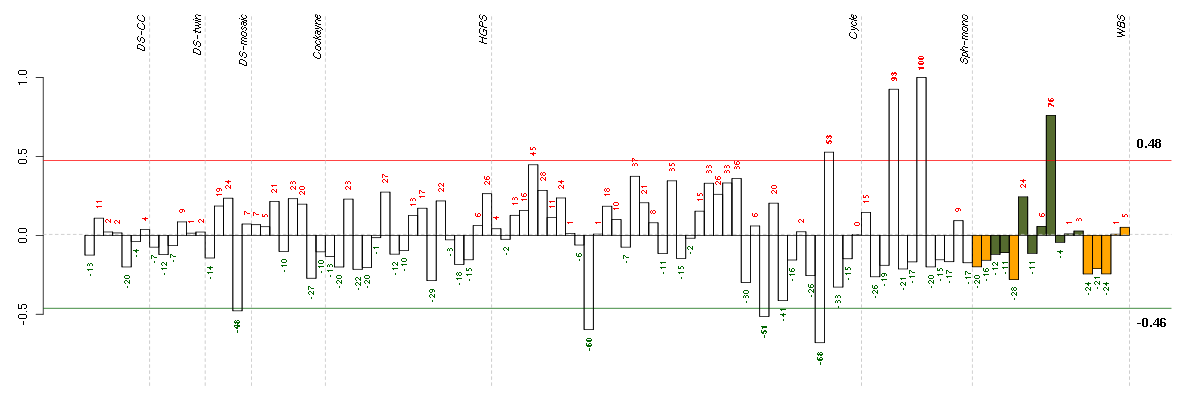
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 6089_CNTL.CEL | 9 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |