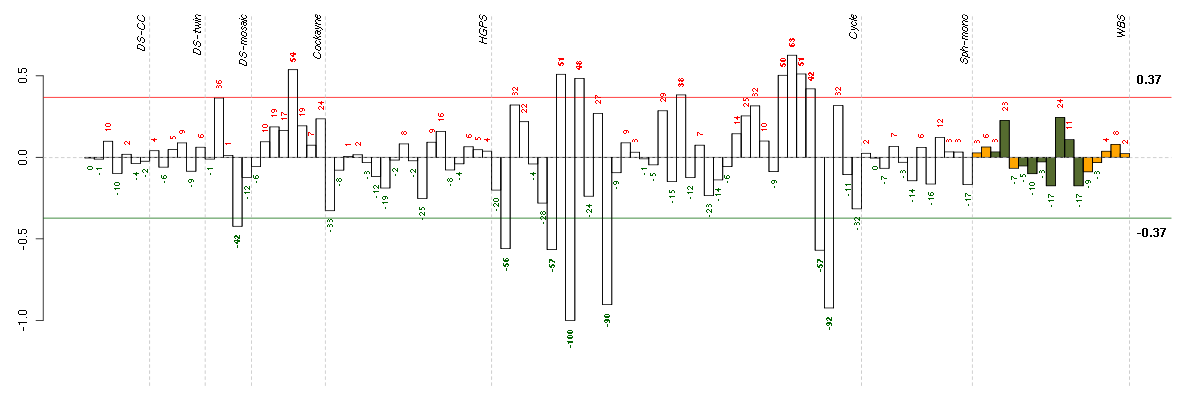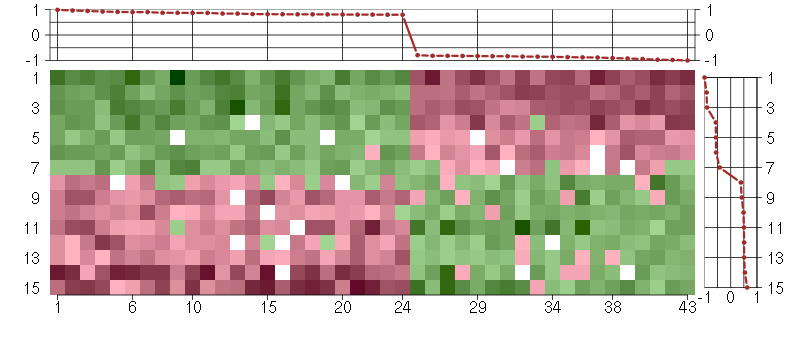

Under-expression is coded with green,
over-expression with red color.



ADORA2Badenosine A2b receptor (205891_at), score: 0.99 AHI1Abelson helper integration site 1 (221569_at), score: -0.85 AIM1absent in melanoma 1 (212543_at), score: 0.84 ANXA10annexin A10 (210143_at), score: -0.88 APOL6apolipoprotein L, 6 (219716_at), score: 0.81 BMP6bone morphogenetic protein 6 (206176_at), score: -0.88 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213348_at), score: 0.82 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.81 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.87 DEPDC6DEP domain containing 6 (218858_at), score: 0.96 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.87 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.82 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.91 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.83 FNBP1formin binding protein 1 (212288_at), score: 0.8 GKglycerol kinase (207387_s_at), score: -0.98 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -1 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.94 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.8 IRF1interferon regulatory factor 1 (202531_at), score: 0.79 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.97 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.82 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.83 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.83 NCALDneurocalcin delta (211685_s_at), score: -0.82 NEFMneurofilament, medium polypeptide (205113_at), score: -0.92 NPTX1neuronal pentraxin I (204684_at), score: -0.85 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.8 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.85 PCDH9protocadherin 9 (219737_s_at), score: -0.94 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.87 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.81 RNF44ring finger protein 44 (203286_at), score: 0.8 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.87 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.82 SMAD3SMAD family member 3 (218284_at), score: 0.9 SNNstannin (218032_at), score: 0.87 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.81 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.91 TLR3toll-like receptor 3 (206271_at), score: 0.92 TMEM2transmembrane protein 2 (218113_at), score: -0.79 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.9 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.84
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |