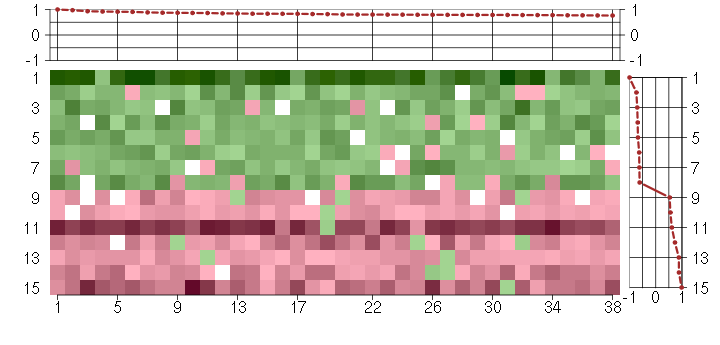

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
sensory perception
The series of events required for an organism to receive a sensory stimulus, convert it to a molecular signal, and recognize and characterize the signal.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
cognition
The operation of the mind by which an organism becomes aware of objects of thought or perception; it includes the mental activities associated with thinking, learning, and memory.
all
This term is the most general term possible


protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
MAP-kinase scaffold activity
Functions as a physical support for the assembly of a multiprotein mitogen-activated protein kinase (MAPK) complex. MAPK scaffold proteins have binding sites for MAPK pathway kinases as well as for upstream signaling proteins.
structural molecule activity
The action of a molecule that contributes to the structural integrity of a complex or assembly within or outside a cell.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
receptor signaling complex scaffold activity
Functions to provide a physical support for the assembly of a multiprotein receptor signaling complex.
protein complex scaffold
Functions to provide a physical support for the assembly of a multiprotein complex.
all
This term is the most general term possible
protein complex scaffold
Functions to provide a physical support for the assembly of a multiprotein complex.
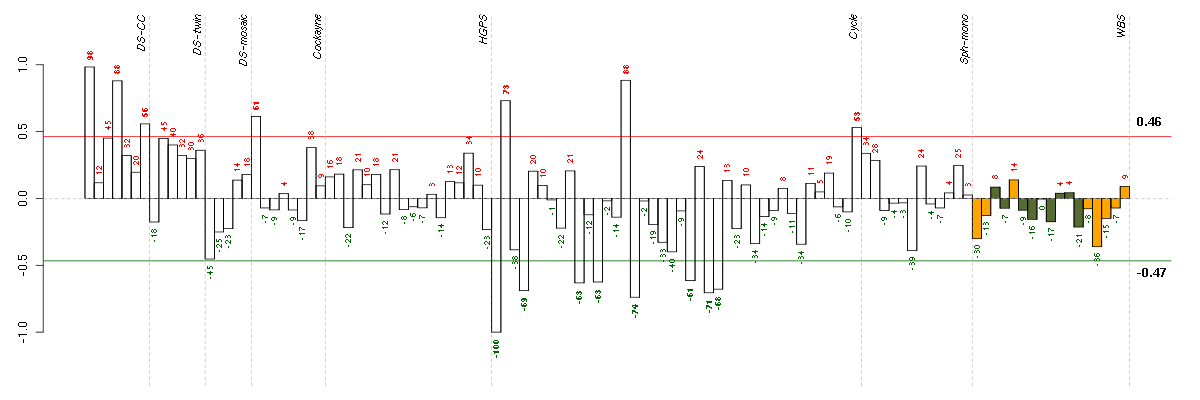
ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.8 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: 0.79 BTNL3butyrophilin-like 3 (217207_s_at), score: 0.8 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.79 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.8 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.87 CYBBcytochrome b-245, beta polypeptide (203923_s_at), score: 0.87 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.79 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.79 EDAectodysplasin A (211130_x_at), score: 0.97 FGL1fibrinogen-like 1 (205305_at), score: 0.77 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.82 HAB1B1 for mucin (215778_x_at), score: 0.92 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: 0.83 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: 0.78 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.77 LOC80054hypothetical LOC80054 (220465_at), score: 0.83 LRRN2leucine rich repeat neuronal 2 (216164_at), score: 0.83 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.8 MAPK8IP2mitogen-activated protein kinase 8 interacting protein 2 (205050_s_at), score: 0.76 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.91 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.89 MYO1Amyosin IA (211916_s_at), score: 0.86 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: 0.78 OR1E2olfactory receptor, family 1, subfamily E, member 2 (208587_s_at), score: 0.79 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: 0.78 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.77 PIPOXpipecolic acid oxidase (221605_s_at), score: 0.84 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: 0.93 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.91 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 0.79 S100A14S100 calcium binding protein A14 (218677_at), score: 0.83 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.85 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.82 TBX21T-box 21 (220684_at), score: 0.8 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 1 TULP2tubby like protein 2 (206733_at), score: 0.79 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.86
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |