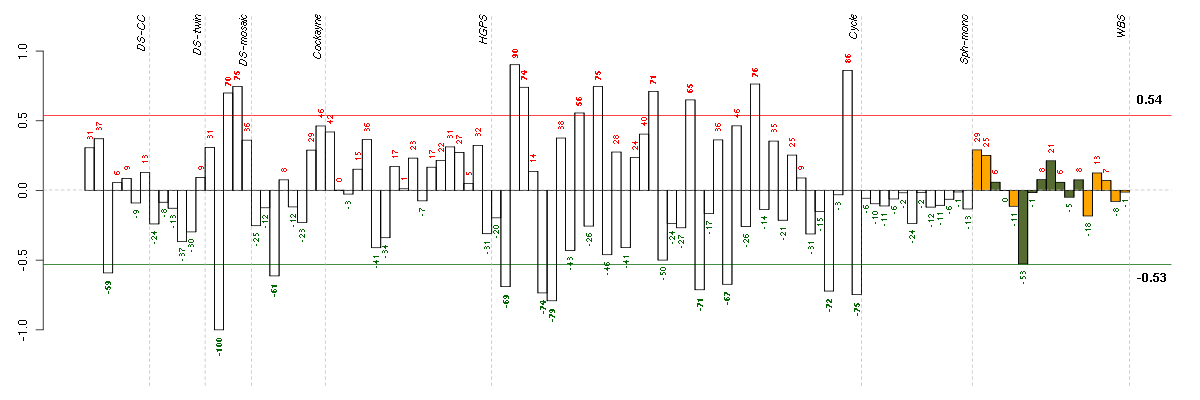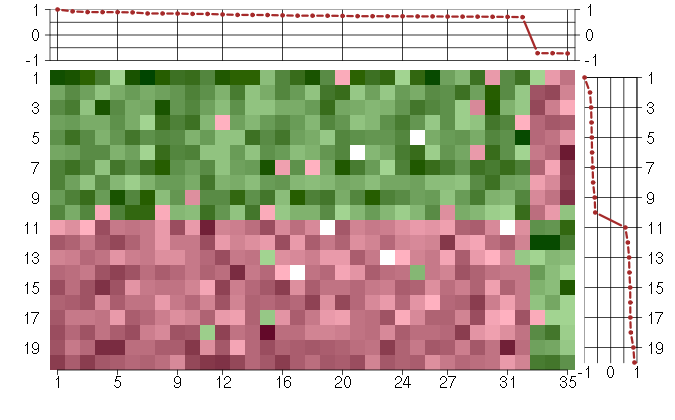

Under-expression is coded with green,
over-expression with red color.



| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04010 | 1.489e-02 | 0.9552 | 6 | 186 | MAPK signaling pathway |
| 05220 | 1.909e-02 | 0.3338 | 4 | 65 | Chronic myeloid leukemia |
| 05212 | 1.978e-02 | 0.339 | 4 | 66 | Pancreatic cancer |
ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.71 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.84 BCL2L1BCL2-like 1 (215037_s_at), score: 0.74 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.84 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.73 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.93 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.7 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.72 FKRPfukutin related protein (219853_at), score: 0.73 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.72 FOXK2forkhead box K2 (203064_s_at), score: 0.71 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.9 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.72 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.77 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.74 NDST1N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 (202608_s_at), score: 0.74 NF1neurofibromin 1 (211094_s_at), score: 0.78 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.79 NUPL2nucleoporin like 2 (204003_s_at), score: -0.72 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.79 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.89 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.76 PXNpaxillin (211823_s_at), score: 1 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.83 RASA3RAS p21 protein activator 3 (206220_s_at), score: 0.76 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.73 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.71 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.71 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.75 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.76 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.84 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.83 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.81 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.9 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.9
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |