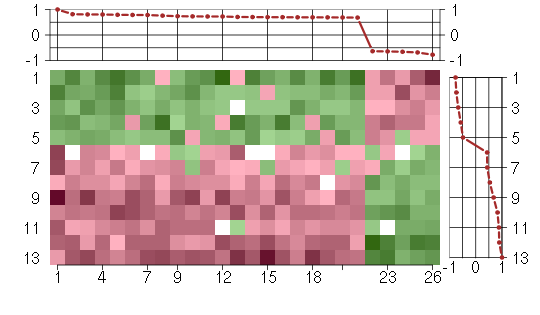

Under-expression is coded with green,
over-expression with red color.



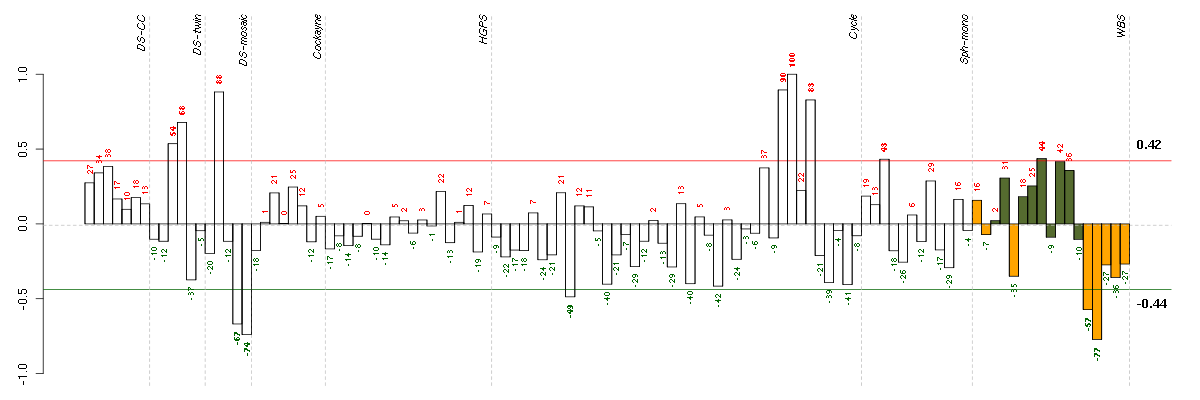
ANGPTL2angiopoietin-like 2 (213001_at), score: 0.69 APOL6apolipoprotein L, 6 (219716_at), score: 0.76 ARHGAP26Rho GTPase activating protein 26 (205068_s_at), score: 0.79 CEBPBCCAAT/enhancer binding protein (C/EBP), beta (212501_at), score: 0.68 CFBcomplement factor B (202357_s_at), score: 0.73 CHST11carbohydrate (chondroitin 4) sulfotransferase 11 (219634_at), score: -0.77 DAPK1death-associated protein kinase 1 (203139_at), score: 0.71 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.81 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.73 FMODfibromodulin (202709_at), score: 0.7 GCH1GTP cyclohydrolase 1 (204224_s_at), score: 0.79 GDF15growth differentiation factor 15 (221577_x_at), score: 0.82 INHBEinhibin, beta E (210587_at), score: 1 IRF1interferon regulatory factor 1 (202531_at), score: 0.74 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.7 MKNK2MAP kinase interacting serine/threonine kinase 2 (218205_s_at), score: 0.69 MOCOSmolybdenum cofactor sulfurase (219959_at), score: 0.8 NFYAnuclear transcription factor Y, alpha (204108_at), score: -0.68 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: -0.64 PPLperiplakin (203407_at), score: 0.7 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.73 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.66 SVEP1sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (213247_at), score: 0.69 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: 0.69 TM4SF1transmembrane 4 L six family member 1 (209386_at), score: -0.64 TRIB3tribbles homolog 3 (Drosophila) (218145_at), score: 0.78
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| D890_WBS.CEL | 13 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 5850_CNTL.CEL | 8 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |