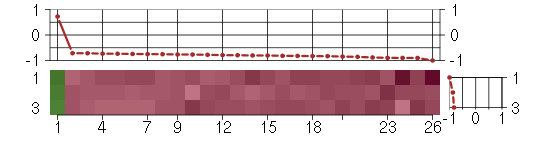

Under-expression is coded with green,
over-expression with red color.



ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.74 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (212224_at), score: -0.86 ARL15ADP-ribosylation factor-like 15 (219842_at), score: -0.8 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: -0.72 COBLL1COBL-like 1 (203642_s_at), score: -0.79 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: -1 FAM46Cfamily with sequence similarity 46, member C (220306_at), score: -0.9 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: -0.83 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: -0.78 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: -0.73 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: -0.85 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: -0.83 GPR126G protein-coupled receptor 126 (213094_at), score: -0.76 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: -0.82 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.88 ISL1ISL LIM homeobox 1 (206104_at), score: -0.71 KCNJ2potassium inwardly-rectifying channel, subfamily J, member 2 (206765_at), score: -0.75 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 0.72 L1CAML1 cell adhesion molecule (204584_at), score: -0.79 LPPR4plasticity related gene 1 (213496_at), score: -0.75 LRRC1leucine rich repeat containing 1 (218816_at), score: -0.73 MANSC1MANSC domain containing 1 (220945_x_at), score: -0.8 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: -0.77 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.82 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: -0.9 TBXA2Rthromboxane A2 receptor (336_at), score: -0.9
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
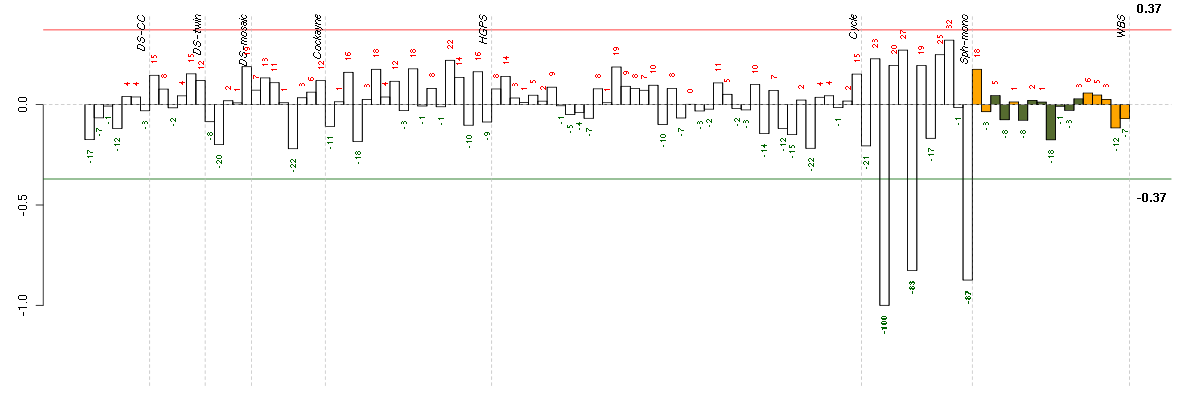
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |