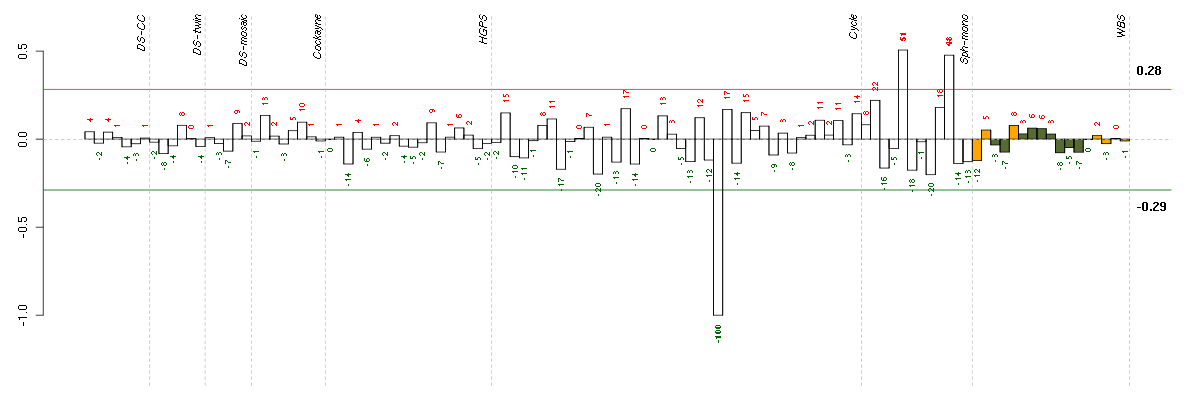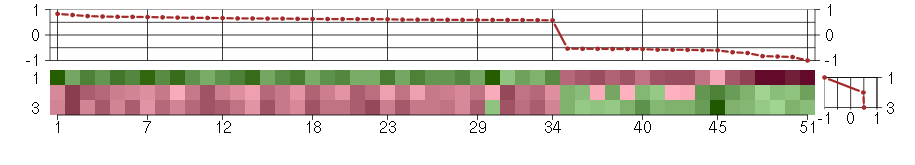

Under-expression is coded with green,
over-expression with red color.



AGAP2ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 (215080_s_at), score: 0.62 AGMATagmatine ureohydrolase (agmatinase) (219792_at), score: -0.86 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.58 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: 0.58 BFSP1beaded filament structural protein 1, filensin (206746_at), score: 0.59 C16orf5chromosome 16 open reading frame 5 (218183_at), score: 0.59 C6orf35chromosome 6 open reading frame 35 (218453_s_at), score: -0.68 C6orf47chromosome 6 open reading frame 47 (204968_at), score: 0.72 CCL8chemokine (C-C motif) ligand 8 (214038_at), score: -0.58 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.63 DKFZp547G183hypothetical LOC55525 (220572_at), score: 0.68 ENGASEendo-beta-N-acetylglucosaminidase (220349_s_at), score: 0.62 FRAT2frequently rearranged in advanced T-cell lymphomas 2 (209864_at), score: -0.55 GRWD1glutamate-rich WD repeat containing 1 (221549_at), score: 0.62 GYG2glycogenin 2 (215695_s_at), score: 0.67 HHIPL2HHIP-like 2 (220283_at), score: 0.66 HIST1H2ALhistone cluster 1, H2al (214554_at), score: -0.84 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.63 HOXA5homeobox A5 (213844_at), score: -0.53 KRT86keratin 86 (215189_at), score: 0.71 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: 0.62 LY75lymphocyte antigen 75 (205668_at), score: -0.71 MAGI1membrane associated guanylate kinase, WW and PDZ domain containing 1 (206144_at), score: -0.83 MAPRE3microtubule-associated protein, RP/EB family, member 3 (203842_s_at), score: 0.67 MGAT4Cmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) (207447_s_at), score: -1 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.65 NECAB3N-terminal EF-hand calcium binding protein 3 (210720_s_at), score: 0.59 NRP2neuropilin 2 (214632_at), score: 0.6 OR1E1olfactory receptor, family 1, subfamily E, member 1 (214515_at), score: -0.58 OR2W1olfactory receptor, family 2, subfamily W, member 1 (221451_s_at), score: -0.55 PJA2praja ring finger 2 (201133_s_at), score: -0.6 PKNOX2PBX/knotted 1 homeobox 2 (63305_at), score: -0.54 PLA2G7phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) (206214_at), score: 0.59 PLCG1phospholipase C, gamma 1 (202789_at), score: 0.64 PSMF1proteasome (prosome, macropain) inhibitor subunit 1 (PI31) (201053_s_at), score: 0.6 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (203169_at), score: 0.64 SARM1sterile alpha and TIR motif containing 1 (213259_s_at), score: 0.74 SHPKsedoheptulokinase (219713_at), score: 0.83 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.6 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: -0.54 STACSH3 and cysteine rich domain (205743_at), score: 0.6 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: 0.63 THG1LtRNA-histidine guanylyltransferase 1-like (S. cerevisiae) (219122_s_at), score: 0.69 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: 0.65 TNFRSF8tumor necrosis factor receptor superfamily, member 8 (206729_at), score: 0.7 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: 0.59 WDR52WD repeat domain 52 (221103_s_at), score: -0.55 WWC1WW and C2 domain containing 1 (213085_s_at), score: 0.78 ZNF394zinc finger protein 394 (214714_at), score: 0.71 ZNF510zinc finger protein 510 (206053_at), score: -0.58 ZNF814zinc finger protein 814 (60794_f_at), score: -0.6
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |