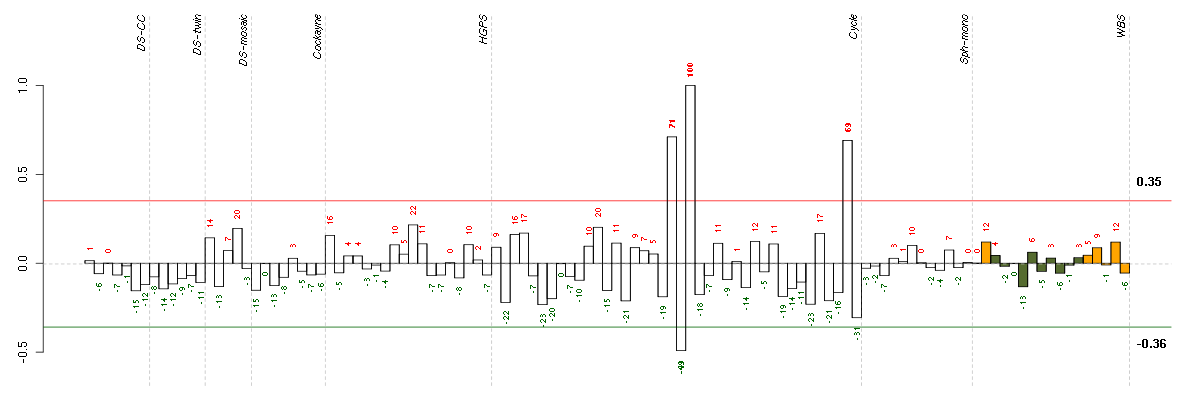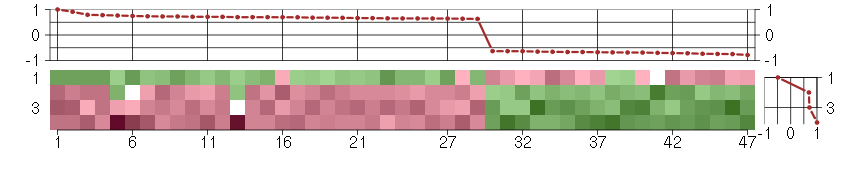

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cell death
The specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
death
A permanent cessation of all vital functions: the end of life; can be applied to a whole organism or to a part of an organism.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
all
This term is the most general term possible
cell death
The specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death.


| Id | Pvalue | ExpCount | Count | Size |
|---|---|---|---|---|
| miR-15/16/195/424/497 | 2.123e-02 | 2.65 | 12 | 633 |
ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.73 ADRB2adrenergic, beta-2-, receptor, surface (206170_at), score: 0.71 ANO1anoctamin 1, calcium activated chloride channel (218804_at), score: 0.68 BAIAP3BAI1-associated protein 3 (216356_x_at), score: -0.71 BCL7AB-cell CLL/lymphoma 7A (203795_s_at), score: 0.7 BHLHE41basic helix-loop-helix family, member e41 (221530_s_at), score: -0.72 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.67 C10orf81chromosome 10 open reading frame 81 (219857_at), score: 0.77 C2CD2LC2CD2-like (204757_s_at), score: 0.63 C8orf17chromosome 8 open reading frame 17 (208266_at), score: -0.67 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: -0.74 CABC1chaperone, ABC1 activity of bc1 complex homolog (S. pombe) (218168_s_at), score: -0.64 CHRNA6cholinergic receptor, nicotinic, alpha 6 (207568_at), score: 0.73 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.72 CRKRSCdc2-related kinase, arginine/serine-rich (219226_at), score: 0.64 EDARectodysplasin A receptor (220048_at), score: -0.69 ENO3enolase 3 (beta, muscle) (204483_at), score: -0.69 ETV7ets variant 7 (221680_s_at), score: -0.63 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.78 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.68 GP6glycoprotein VI (platelet) (220336_s_at), score: 0.7 GPR157G protein-coupled receptor 157 (220901_at), score: 0.75 HTN1histatin 1 (206639_x_at), score: -0.66 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.66 KCTD7potassium channel tetramerisation domain containing 7 (213474_at), score: -0.68 KRT33Akeratin 33A (208483_x_at), score: 0.64 KRTAP1-1keratin associated protein 1-1 (220976_s_at), score: 0.7 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.65 LRRC37A2leucine rich repeat containing 37, member A2 (221740_x_at), score: -0.68 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.7 MBNL2muscleblind-like 2 (Drosophila) (205018_s_at), score: -0.65 MNTMAX binding protein (204206_at), score: -0.7 NR1D2nuclear receptor subfamily 1, group D, member 2 (209750_at), score: -0.78 PLAGL2pleiomorphic adenoma gene-like 2 (202925_s_at), score: 0.66 PRKCDprotein kinase C, delta (202545_at), score: 1 PTPRRprotein tyrosine phosphatase, receptor type, R (210675_s_at), score: 0.79 RASSF9Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 (210335_at), score: -0.74 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: -0.64 RRP8ribosomal RNA processing 8, methyltransferase, homolog (yeast) (203171_s_at), score: 0.71 TAOK2TAO kinase 2 (204986_s_at), score: 0.73 TBPTATA box binding protein (203135_at), score: 0.67 TBX21T-box 21 (220684_at), score: -0.75 ZDHHC13zinc finger, DHHC-type containing 13 (219296_at), score: 0.65 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: 0.91 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.65 ZNF318zinc finger protein 318 (203521_s_at), score: 0.69 ZNF443zinc finger protein 443 (205928_at), score: 0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |