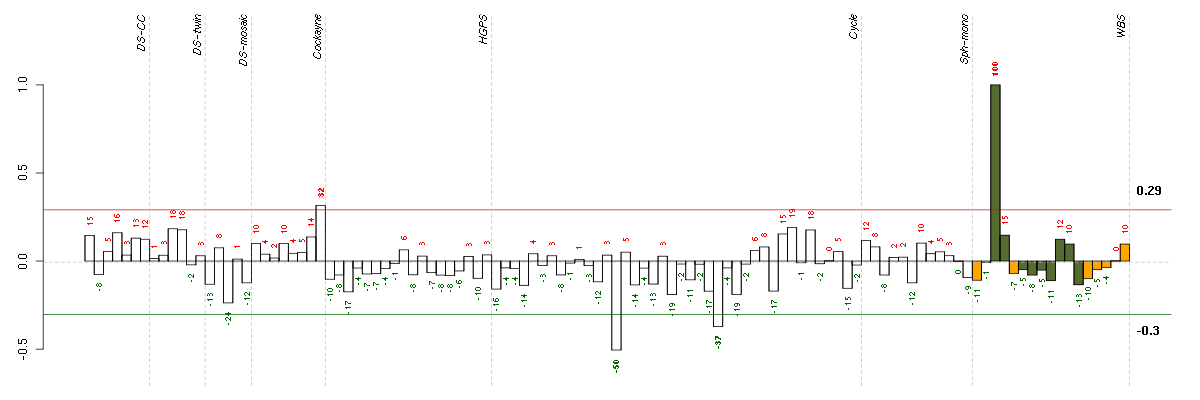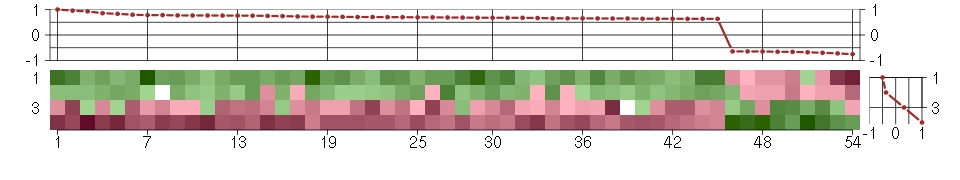

Under-expression is coded with green,
over-expression with red color.



ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.93 AMTaminomethyltransferase (204294_at), score: 0.72 APOL3apolipoprotein L, 3 (221087_s_at), score: 0.76 ATMataxia telangiectasia mutated (210858_x_at), score: 0.65 ATXN1ataxin 1 (203231_s_at), score: 0.68 AVILadvillin (205539_at), score: 0.78 C1QTNF3C1q and tumor necrosis factor related protein 3 (220988_s_at), score: 0.83 CDO1cysteine dioxygenase, type I (204154_at), score: 0.69 CLN3ceroid-lipofuscinosis, neuronal 3 (209275_s_at), score: 0.65 COL16A1collagen, type XVI, alpha 1 (204345_at), score: 0.7 COL21A1collagen, type XXI, alpha 1 (208096_s_at), score: 0.77 CROCCciliary rootlet coiled-coil, rootletin (206274_s_at), score: 0.78 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: 0.7 EPHB3EPH receptor B3 (1438_at), score: 0.75 GPATCH3G patch domain containing 3 (218895_at), score: 0.74 GZMMgranzyme M (lymphocyte met-ase 1) (207460_at), score: 0.67 HEBP2heme binding protein 2 (203430_at), score: -0.68 HIST2H2AA3histone cluster 2, H2aa3 (214290_s_at), score: -0.7 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.69 IFI27interferon, alpha-inducible protein 27 (202411_at), score: 0.67 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: 0.69 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.64 KIAA0513KIAA0513 (204546_at), score: 0.65 MARCH5membrane-associated ring finger (C3HC4) 5 (218582_at), score: -0.65 MCM3APASMCM3AP antisense RNA (non-protein coding) (220459_at), score: -0.72 MDKmidkine (neurite growth-promoting factor 2) (209035_at), score: 0.64 MGLLmonoglyceride lipase (211026_s_at), score: -0.66 MMP12matrix metallopeptidase 12 (macrophage elastase) (204580_at), score: -0.66 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.64 NRP2neuropilin 2 (214632_at), score: 0.76 OGG18-oxoguanine DNA glycosylase (205760_s_at), score: 0.64 OR2S2olfactory receptor, family 2, subfamily S, member 2 (221409_at), score: -0.75 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: 0.63 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: 0.64 PTCRApre T-cell antigen receptor alpha (211252_x_at), score: 0.65 PVT1Pvt1 oncogene (non-protein coding) (216249_at), score: -0.64 RAB26RAB26, member RAS oncogene family (50965_at), score: 0.68 RAB30RAB30, member RAS oncogene family (206530_at), score: 0.72 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: 0.65 RBKSribokinase (219222_at), score: 0.77 RNFT1ring finger protein, transmembrane 1 (221194_s_at), score: 0.8 ROGDIrogdi homolog (Drosophila) (218394_at), score: 0.73 RP4-691N24.1ninein-like (207705_s_at), score: 0.76 RP5-1077B9.4invasion inhibitory protein 45 (48659_at), score: 0.71 SAMD14sterile alpha motif domain containing 14 (213866_at), score: 0.68 SIM2single-minded homolog 2 (Drosophila) (206558_at), score: 0.77 SMA5glucuronidase, beta pseudogene (215043_s_at), score: 1 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: 0.63 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.7 TROtrophinin (211700_s_at), score: 0.96 UBIAD1UbiA prenyltransferase domain containing 1 (219131_at), score: 0.65 ZNF248zinc finger protein 248 (213269_at), score: 0.71 ZNF669zinc finger protein 669 (220215_at), score: 0.68 ZSCAN12zinc finger and SCAN domain containing 12 (206507_at), score: 0.85
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| 1104_CNTL.CEL | 3 | 8 | WBS | hgu133plus2 | none | WBS 1 |