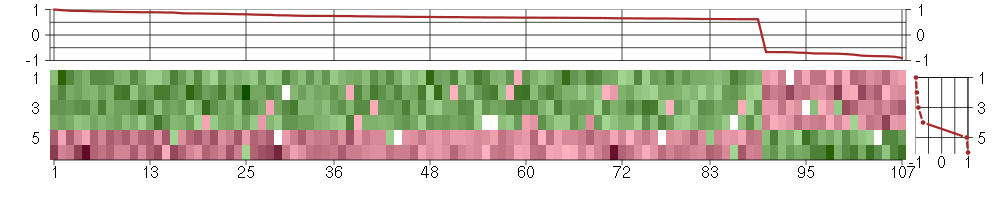

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
defense response to bacterium
Reactions triggered in response to the presence of a bacterium that act to protect the cell or organism.
vesicle-mediated transport
The directed movement of substances, either within a vesicle or in the vesicle membrane, into, out of or within a cell.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism.
endocytosis
A type of vesicle-mediated transport in which cells take up external materials or membrane constituents by the invagination of small region of the plasma membrane to form a new membrane-bounded vesicle.
phagocytosis
The process whereby phagocytes engulf external particulate material. The particles are initially contained within phagocytic vacuoles (phagosomes), which then fuse with primary lysosomes to effect digestion of the particles.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to biotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a biotic stimulus, a stimulus caused or produced by a living organism.
response to other organism
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism.
response to bacterium
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a bacterium.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
membrane invagination
The infolding of a membrane, resulting in formation of a vesicle.
cellular component organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a cellular component.
membrane organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a membrane. A membrane is a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
multi-organism process
Any process by which an organism has an effect on another organism of the same or different species.
all
This term is the most general term possible
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
response to other organism
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism.
vesicle-mediated transport
The directed movement of substances, either within a vesicle or in the vesicle membrane, into, out of or within a cell.
defense response to bacterium
Reactions triggered in response to the presence of a bacterium that act to protect the cell or organism.
endocytosis
A type of vesicle-mediated transport in which cells take up external materials or membrane constituents by the invagination of small region of the plasma membrane to form a new membrane-bounded vesicle.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

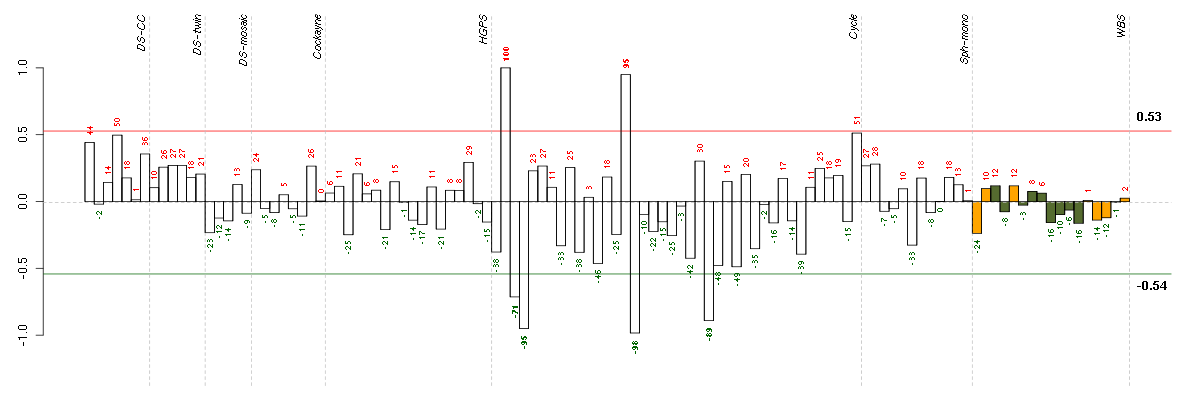
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.94 ADORA1adenosine A1 receptor (216220_s_at), score: 0.75 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: 0.66 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.7 ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: 0.69 APOC4apolipoprotein C-IV (206738_at), score: 0.63 APPamyloid beta (A4) precursor protein (214953_s_at), score: 0.89 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.77 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.82 BEST1bestrophin 1 (207671_s_at), score: 0.64 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: -0.75 C1orf69chromosome 1 open reading frame 69 (215490_at), score: 0.7 C2orf37chromosome 2 open reading frame 37 (220172_at), score: -0.72 C4Acomplement component 4A (Rodgers blood group) (214428_x_at), score: 0.71 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.67 CATSPER2cation channel, sperm associated 2 (217588_at), score: 0.65 CCDC33coiled-coil domain containing 33 (220908_at), score: 0.83 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.91 CCDC94coiled-coil domain containing 94 (204335_at), score: 0.64 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.79 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: 0.75 CNNM3cyclin M3 (220739_s_at), score: -0.67 CYorf14chromosome Y open reading frame 14 (207063_at), score: -0.83 CYTH4cytohesin 4 (219183_s_at), score: 0.76 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: 0.63 DLX4distal-less homeobox 4 (208216_at), score: 0.62 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.88 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.85 EDAectodysplasin A (211130_x_at), score: 0.89 EPN1epsin 1 (221141_x_at), score: 0.77 ETNK2ethanolamine kinase 2 (219268_at), score: 0.63 F11coagulation factor XI (206610_s_at), score: 0.67 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: 0.62 FGL1fibrinogen-like 1 (205305_at), score: 0.68 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 1 FLCNfolliculin (215645_at), score: 0.67 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: -0.81 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.89 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: 0.73 GPR144G protein-coupled receptor 144 (216289_at), score: 0.97 GPR32G protein-coupled receptor 32 (221469_at), score: 0.81 GRM1glutamate receptor, metabotropic 1 (210939_s_at), score: 0.68 HAB1B1 for mucin (215778_x_at), score: 0.91 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.72 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: 0.65 HIST1H2BChistone cluster 1, H2bc (214455_at), score: -0.69 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: 0.65 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: 0.73 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.72 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.9 KIF26Bkinesin family member 26B (220002_at), score: 0.72 KRT8keratin 8 (209008_x_at), score: 0.69 KRT8P12keratin 8 pseudogene 12 (222060_at), score: 0.66 KSR1kinase suppressor of ras 1 (213769_at), score: 0.65 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: 0.83 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.81 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.72 LOC149501similar to keratin 8 (216821_at), score: 0.62 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.68 MAPK8IP2mitogen-activated protein kinase 8 interacting protein 2 (205050_s_at), score: 0.62 MERTKc-mer proto-oncogene tyrosine kinase (211913_s_at), score: 0.65 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.69 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.74 MYO15Amyosin XVA (220288_at), score: 0.67 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.67 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.75 OPRL1opiate receptor-like 1 (206564_at), score: 0.85 PCNXL2pecanex-like 2 (Drosophila) (39650_s_at), score: 0.69 PHF2PHD finger protein 2 (212726_at), score: -0.73 PIPOXpipecolic acid oxidase (221605_s_at), score: 0.67 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.95 PLXNB3plexin B3 (205957_at), score: 0.71 PMS2L2postmeiotic segregation increased 2-like 2 pseudogene (215410_at), score: 0.9 PNMAL1PNMA-like 1 (218824_at), score: -0.83 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.84 PRLHprolactin releasing hormone (221443_x_at), score: 0.68 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.85 RBM12BRNA binding motif protein 12B (51228_at), score: -0.85 RBM38RNA binding motif protein 38 (212430_at), score: 0.79 REG3Aregenerating islet-derived 3 alpha (205815_at), score: 0.65 RLN1relaxin 1 (211753_s_at), score: 0.74 RNF121ring finger protein 121 (219021_at), score: 0.92 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.67 SEPT5septin 5 (209767_s_at), score: 0.68 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.63 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: -0.9 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.71 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: 0.73 SNTG1syntrophin, gamma 1 (220405_at), score: -0.73 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: 0.68 SPATA1spermatogenesis associated 1 (221057_at), score: 0.7 SSX3synovial sarcoma, X breakpoint 3 (211731_x_at), score: 0.71 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.93 STIM1stromal interaction molecule 1 (202764_at), score: -0.7 SYT1synaptotagmin I (203999_at), score: -0.67 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.8 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.69 TGM4transglutaminase 4 (prostate) (217566_s_at), score: 0.68 TMPRSS6transmembrane protease, serine 6 (214955_at), score: 0.94 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.75 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.75 TP53TG1TP53 target 1 (non-protein coding) (210241_s_at), score: 0.62 TULP2tubby like protein 2 (206733_at), score: 0.88 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.64 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.7 ZNF192zinc finger protein 192 (206579_at), score: -0.82 ZNF214zinc finger protein 214 (220497_at), score: -0.77
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |