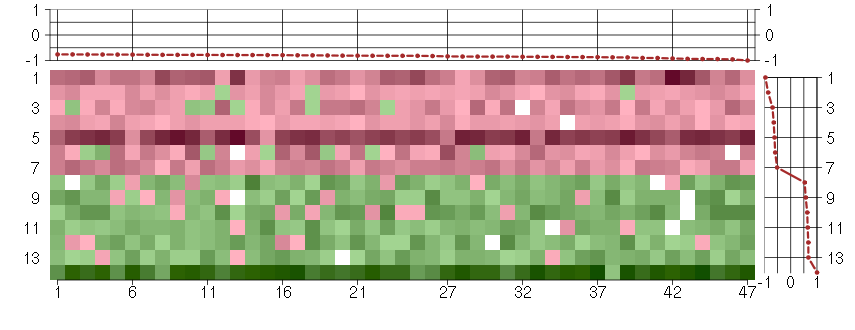

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

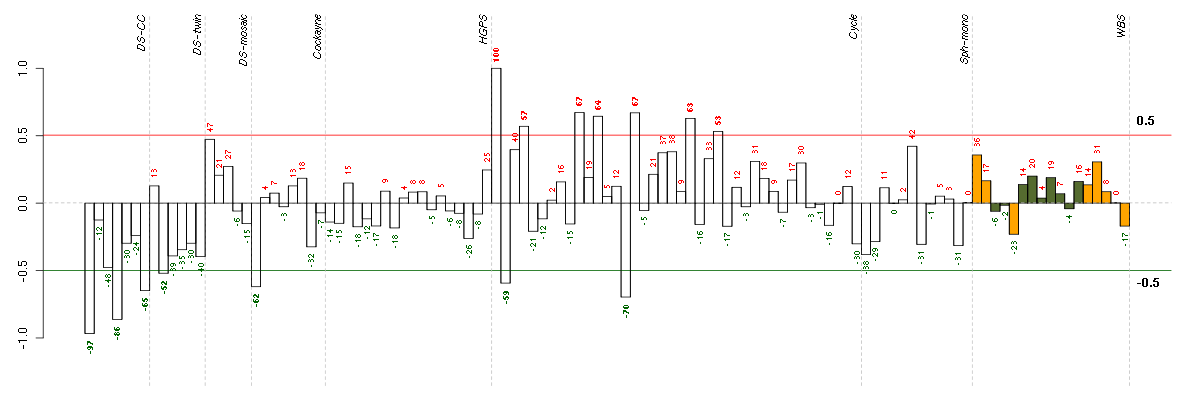
A1CFAPOBEC1 complementation factor (220951_s_at), score: -0.82 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: -0.84 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: -0.78 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.9 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.82 C8orf60chromosome 8 open reading frame 60 (220712_at), score: -0.85 CCDC9coiled-coil domain containing 9 (206257_at), score: -0.82 CD53CD53 molecule (203416_at), score: -0.77 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.94 CYBBcytochrome b-245, beta polypeptide (203923_s_at), score: -0.81 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.79 DHRS9dehydrogenase/reductase (SDR family) member 9 (219799_s_at), score: -0.78 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: -0.8 DRD5dopamine receptor D5 (208486_at), score: -0.78 EDAectodysplasin A (211130_x_at), score: -0.86 FGL1fibrinogen-like 1 (205305_at), score: -0.83 HAB1B1 for mucin (215778_x_at), score: -0.88 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.85 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.76 KCNQ3potassium voltage-gated channel, KQT-like subfamily, member 3 (206573_at), score: -0.77 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: -0.87 LOC80054hypothetical LOC80054 (220465_at), score: -0.86 LRRN2leucine rich repeat neuronal 2 (216164_at), score: -0.78 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: -0.79 MMRN2multimerin 2 (219091_s_at), score: -0.79 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.94 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.87 MYO1Amyosin IA (211916_s_at), score: -0.88 NCRNA00092non-protein coding RNA 92 (215861_at), score: -0.79 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: -0.8 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: -0.76 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -0.76 PGCprogastricsin (pepsinogen C) (205261_at), score: -0.81 PGK2phosphoglycerate kinase 2 (217009_at), score: -0.78 PIPOXpipecolic acid oxidase (221605_s_at), score: -0.85 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: -0.93 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: -0.96 RLN1relaxin 1 (211753_s_at), score: -0.78 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: -0.77 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -0.85 S100A14S100 calcium binding protein A14 (218677_at), score: -0.9 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: -0.8 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.85 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: -0.81 TBX21T-box 21 (220684_at), score: -0.86 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -1 VGFVGF nerve growth factor inducible (205586_x_at), score: -0.92
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |