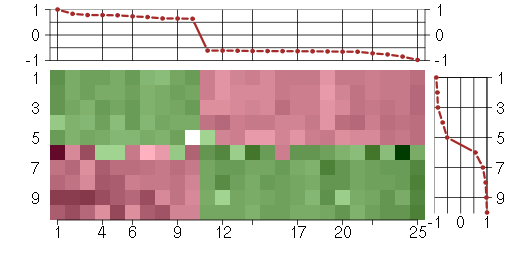

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
collagen
Any of the various assemblies in which collagen chains form a left-handed triple helix; may assemble into higher order structures.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
fibrillar collagen
Any collagen polymer in which collagen triple helices associate to form fibrils.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).
collagen
Any of the various assemblies in which collagen chains form a left-handed triple helix; may assemble into higher order structures.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
growth factor binding
Interacting selectively with any growth factor, proteins or polypeptides that stimulate a cell or organism to grow or proliferate.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
platelet-derived growth factor binding
Interacting selectively with platelet-derived growth factor.
all
This term is the most general term possible
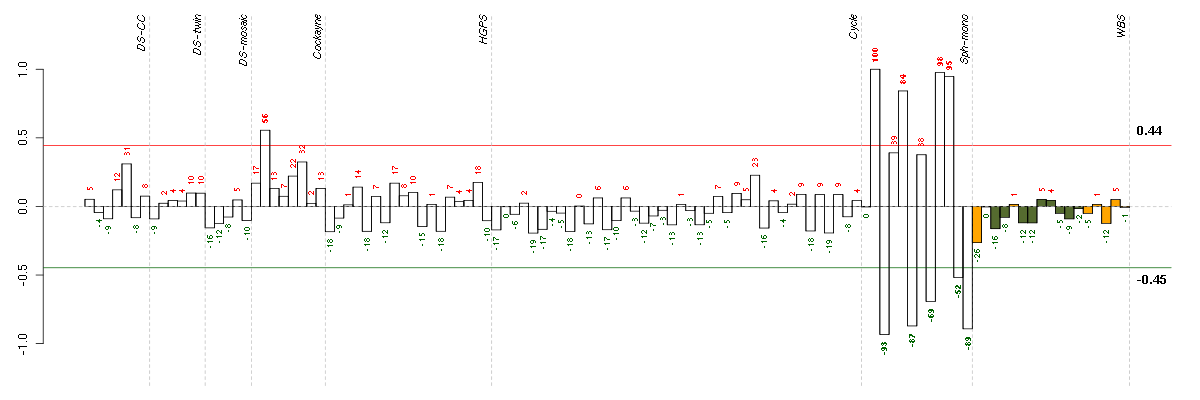
AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.83 C14orf169chromosome 14 open reading frame 169 (219526_at), score: -0.63 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: 0.7 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: 0.73 COL3A1collagen, type III, alpha 1 (215076_s_at), score: -0.63 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.64 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.65 GALCgalactosylceramidase (204417_at), score: -0.61 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: -0.98 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: 0.78 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 1 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: 0.78 LUMlumican (201744_s_at), score: -0.85 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: 0.77 MYO1Bmyosin IB (212364_at), score: -0.76 PCTK2PCTAIRE protein kinase 2 (221918_at), score: -0.61 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: -0.65 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: -0.66 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: 0.65 SPANXCSPANX family, member C (220217_x_at), score: 0.65 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: -0.62 TEStestis derived transcript (3 LIM domains) (202720_at), score: -0.72 THBS2thrombospondin 2 (203083_at), score: -0.64 TNS1tensin 1 (221748_s_at), score: -0.63 TSPYL5TSPY-like 5 (213122_at), score: -0.64
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |