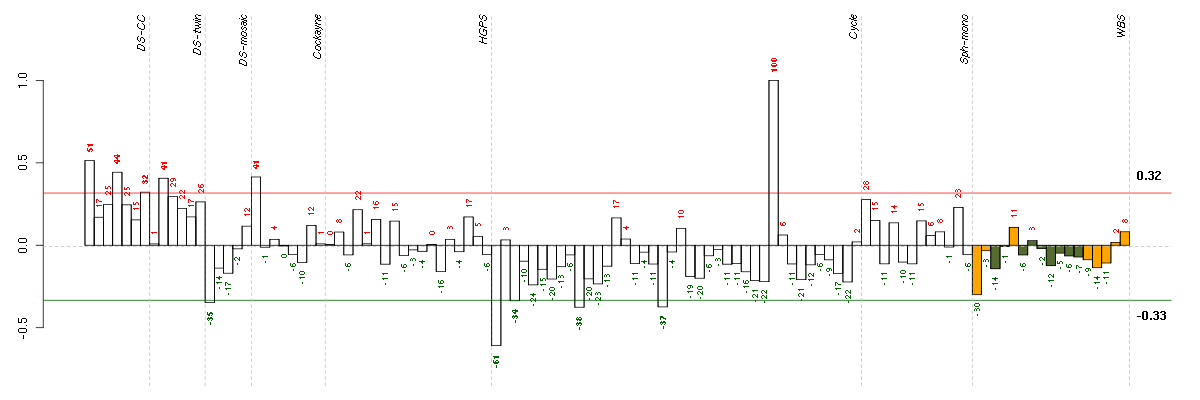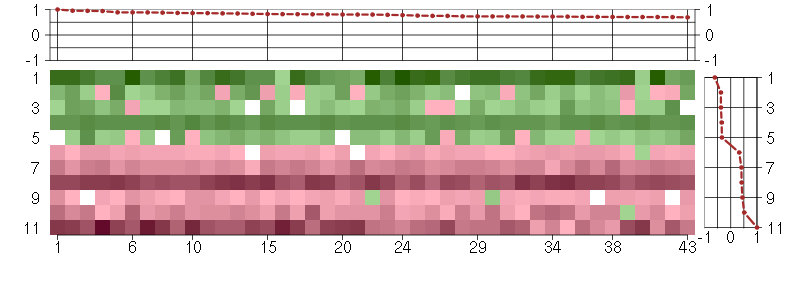

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
transmembrane transporter activity
Catalysis of the transfer of a substance from one side of a membrane to the other.
organic acid transmembrane transporter activity
Catalysis of the transfer of organic acids, any acidic compound containing carbon in covalent linkage, from one side of the membrane to the other.
monocarboxylic acid transmembrane transporter activity
Catalysis of the transfer of monocarboxylic acids from one side of the membrane to the other. A monocarboxylic acid is an organic acid with one COOH group.
bile acid transmembrane transporter activity
Catalysis of the transfer of bile acid from one side of the membrane to the other. Bile acids are any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
substrate-specific transmembrane transporter activity
Catalysis of the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
carboxylic acid transmembrane transporter activity
Catalysis of the transfer of carboxylic acids from one side of the membrane to the other. Carboxylic acids are organic acids containing one or more carboxyl (COOH) groups or anions (COO-).
all
This term is the most general term possible
substrate-specific transmembrane transporter activity
Catalysis of the transfer of a specific substance or group of related substances from one side of a membrane to the other.
ADRBK2adrenergic, beta, receptor kinase 2 (204184_s_at), score: 0.82 AFF2AF4/FMR2 family, member 2 (206105_at), score: 0.89 ATRNL1attractin-like 1 (213744_at), score: 0.82 CA1carbonic anhydrase I (205949_at), score: 0.94 CD53CD53 molecule (203416_at), score: 0.85 CLCA2chloride channel accessory 2 (217528_at), score: 0.83 CLDN10claudin 10 (205328_at), score: 0.81 CLEC2DC-type lectin domain family 2, member D (220132_s_at), score: 0.73 CNKSR2connector enhancer of kinase suppressor of Ras 2 (206731_at), score: 0.95 CTRLchymotrypsin-like (214377_s_at), score: 0.89 DAOD-amino-acid oxidase (206878_at), score: 0.76 DSC2desmocollin 2 (204751_x_at), score: 0.75 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: 0.87 IL16interleukin 16 (lymphocyte chemoattractant factor) (209827_s_at), score: 0.73 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (220663_at), score: 0.75 KRT2keratin 2 (207908_at), score: 0.95 LHX3LIM homeobox 3 (221670_s_at), score: 0.72 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: 0.72 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: 0.84 MBmyoglobin (204179_at), score: 0.71 MIA2melanoma inhibitory activity 2 (221177_at), score: 0.73 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: 0.79 NR0B1nuclear receptor subfamily 0, group B, member 1 (206644_at), score: 0.82 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.8 OR2W1olfactory receptor, family 2, subfamily W, member 1 (221451_s_at), score: 0.69 PAX4paired box 4 (207867_at), score: 0.73 PEG3paternally expressed 3 (209242_at), score: 0.71 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.73 PGK2phosphoglycerate kinase 2 (217009_at), score: 0.72 PLLPplasma membrane proteolipid (plasmolipin) (204519_s_at), score: 0.7 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: 0.8 RAPGEF4Rap guanine nucleotide exchange factor (GEF) 4 (205651_x_at), score: 0.88 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 1 SIRPB1signal-regulatory protein beta 1 (206934_at), score: 0.86 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (207095_at), score: 0.7 SLC26A4solute carrier family 26, member 4 (206529_x_at), score: 0.86 SLC28A3solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 (220475_at), score: 0.7 SLCO1A2solute carrier organic anion transporter family, member 1A2 (207308_at), score: 0.8 TAAR2trace amine associated receptor 2 (221394_at), score: 0.71 TACR1tachykinin receptor 1 (208048_at), score: 0.89 TMEM19transmembrane protein 19 (219941_at), score: 0.7 TP63tumor protein p63 (209863_s_at), score: 0.78 ZNF335zinc finger protein 335 (78330_at), score: 0.81
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |