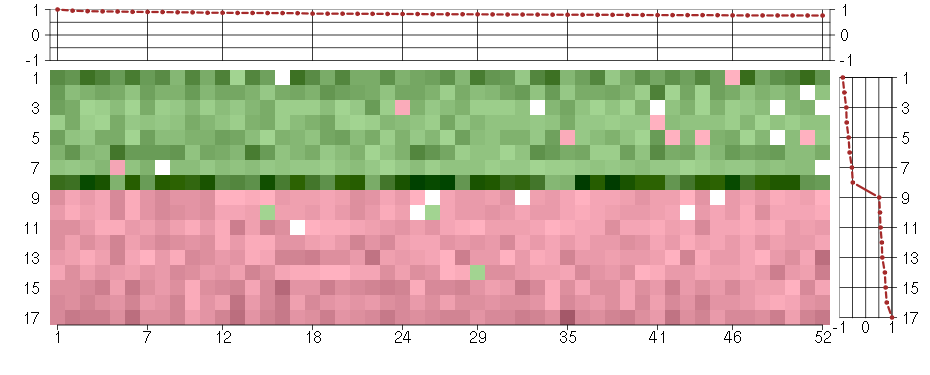

Under-expression is coded with green,
over-expression with red color.


cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

ABI1abl-interactor 1 (209028_s_at), score: 0.78 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.79 ATF1activating transcription factor 1 (222103_at), score: 0.89 ATF2activating transcription factor 2 (212984_at), score: 0.77 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.82 AZI25-azacytidine induced 2 (218043_s_at), score: 0.76 BBS10Bardet-Biedl syndrome 10 (219487_at), score: 0.79 C10orf97chromosome 10 open reading frame 97 (218297_at), score: 0.79 C5orf44chromosome 5 open reading frame 44 (218674_at), score: 0.83 C6orf211chromosome 6 open reading frame 211 (218195_at), score: 0.9 C9orf82chromosome 9 open reading frame 82 (219276_x_at), score: 0.77 CAPZA2capping protein (actin filament) muscle Z-line, alpha 2 (201237_at), score: 0.81 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: 0.77 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.84 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: 0.8 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.9 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.86 FNDC3Afibronectin type III domain containing 3A (202304_at), score: 0.83 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 0.87 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: 0.93 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.86 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (210970_s_at), score: 0.8 IMPACTImpact homolog (mouse) (218637_at), score: 0.81 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.87 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: 0.78 MATR3matrin 3 (200626_s_at), score: 0.77 MED23mediator complex subunit 23 (218846_at), score: 0.87 MYO1Bmyosin IB (212364_at), score: 0.92 MYO6myosin VI (203216_s_at), score: 0.77 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.81 PJA2praja ring finger 2 (201133_s_at), score: 0.78 RAB1ARAB1A, member RAS oncogene family (213440_at), score: 0.78 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: 0.8 SENP6SUMO1/sentrin specific peptidase 6 (202318_s_at), score: 0.8 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.86 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.91 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: 0.78 SPG20spastic paraplegia 20 (Troyer syndrome) (212526_at), score: 0.79 STAM2signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 (209649_at), score: 0.83 STRN3striatin, calmodulin binding protein 3 (204496_at), score: 0.86 STXBP3syntaxin binding protein 3 (203310_at), score: 0.79 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: 0.81 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.95 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: 0.78 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: 0.81 TMEM123transmembrane protein 123 (211967_at), score: 0.82 TMEM30Atransmembrane protein 30A (217743_s_at), score: 1 TMF1TATA element modulatory factor 1 (213024_at), score: 0.93 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: 0.84 YTHDC2YTH domain containing 2 (213077_at), score: 0.89 ZC3H14zinc finger CCCH-type containing 14 (213064_at), score: 0.77 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: 0.84
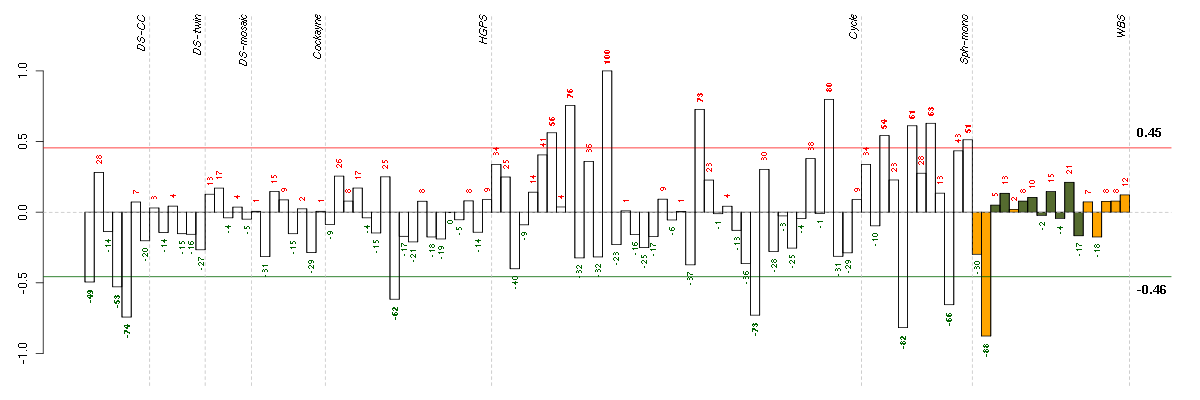
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |