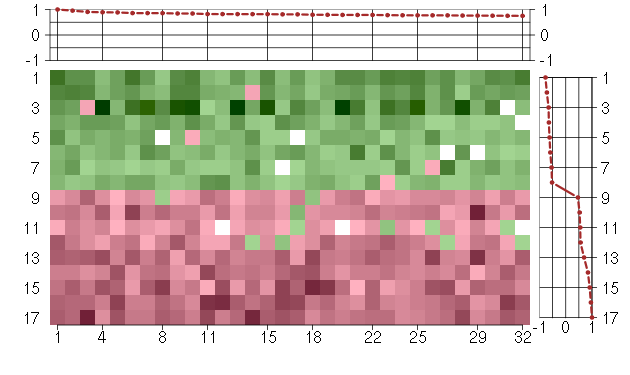

Under-expression is coded with green,
over-expression with red color.



ABI1abl-interactor 1 (209028_s_at), score: 0.77 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.82 AHI1Abelson helper integration site 1 (221569_at), score: 0.81 CCNT2cyclin T2 (204645_at), score: 0.78 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 1 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: 0.86 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.89 EDNRAendothelin receptor type A (204464_s_at), score: 0.79 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: 0.75 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (219532_at), score: 0.77 FNDC3Afibronectin type III domain containing 3A (202304_at), score: 0.77 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: 0.84 GKglycerol kinase (207387_s_at), score: 0.81 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.84 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.82 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: 0.79 NOX4NADPH oxidase 4 (219773_at), score: 0.82 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.95 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: 0.79 SLC2A3P1solute carrier family 2 (facilitated glucose transporter), member 3 pseudogene 1 (221751_at), score: 0.86 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.75 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: 0.79 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.76 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: 0.76 STK38Lserine/threonine kinase 38 like (212572_at), score: 0.76 STRN3striatin, calmodulin binding protein 3 (204496_at), score: 0.79 TMF1TATA element modulatory factor 1 (213024_at), score: 0.82 UFM1ubiquitin-fold modifier 1 (218050_at), score: 0.82 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: 0.86 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: 0.9 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: 0.77 ZFYVE16zinc finger, FYVE domain containing 16 (203651_at), score: 0.89
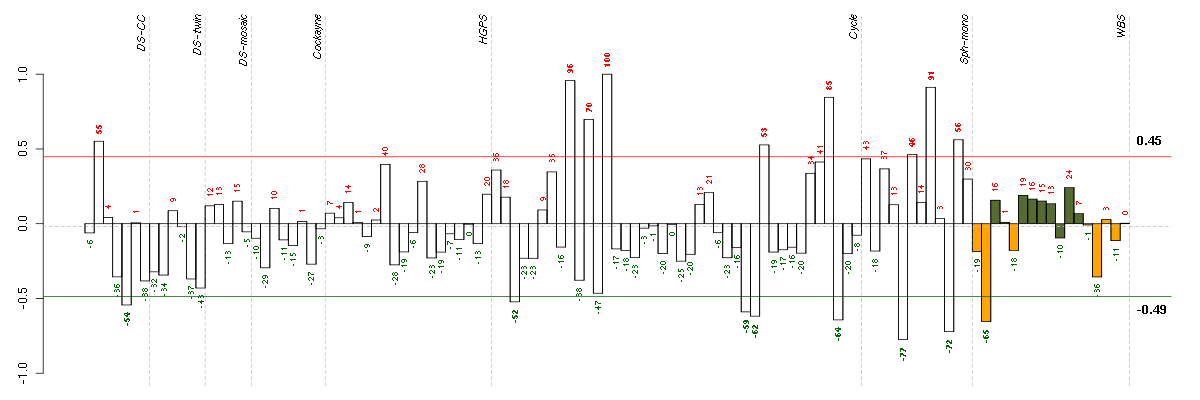
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |