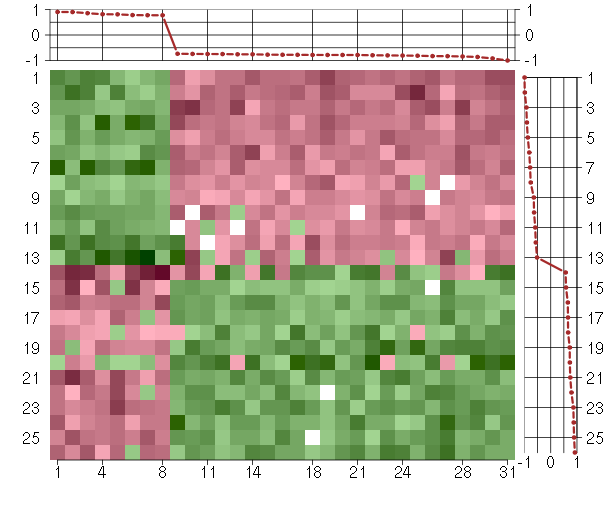

Under-expression is coded with green,
over-expression with red color.


cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
dendrite
A branching protoplasmic process of a neuron that receive and integrate signals coming from axons of other neurons, and convey the resulting signal to the body of the cell.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
neuron projection
A prolongation or process extending from a nerve cell, e.g. an axon or dendrite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

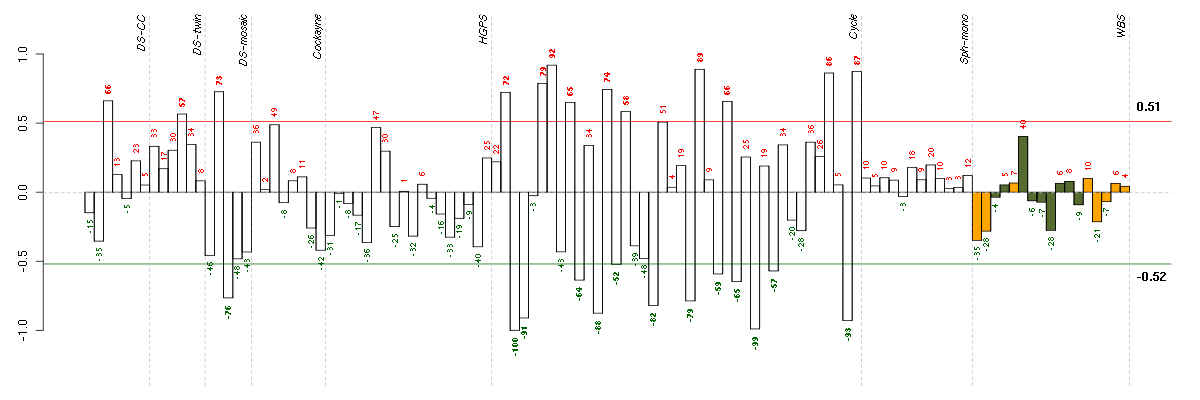
ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.74 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: -0.79 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.78 BCL2L1BCL2-like 1 (215037_s_at), score: -0.78 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.75 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.78 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.86 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.78 FBXO2F-box protein 2 (219305_x_at), score: 0.78 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: -0.81 FOXK2forkhead box K2 (203064_s_at), score: -0.83 HAB1B1 for mucin (215778_x_at), score: 0.81 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.92 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.81 LOC391132similar to hCG2041276 (216177_at), score: 0.9 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: -0.77 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.76 NF1neurofibromin 1 (211094_s_at), score: -0.81 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.76 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: -0.75 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: -0.78 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.78 PXNpaxillin (211823_s_at), score: -1 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.9 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.86 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.8 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.83 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.78 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: 0.77 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.74 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.84
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |