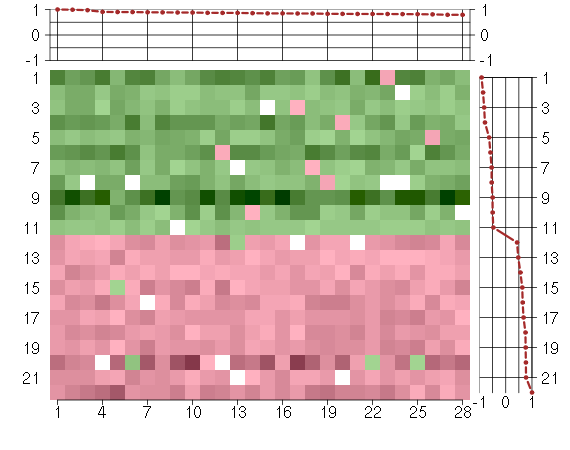

Under-expression is coded with green,
over-expression with red color.



ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (212476_at), score: 1 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.9 BNIP2BCL2/adenovirus E1B 19kDa interacting protein 2 (209308_s_at), score: 0.82 C5orf28chromosome 5 open reading frame 28 (219029_at), score: 0.82 EPS15epidermal growth factor receptor pathway substrate 15 (217887_s_at), score: 0.85 EVI5ecotropic viral integration site 5 (209717_at), score: 0.88 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.99 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.9 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.87 KIAA0562KIAA0562 (204075_s_at), score: 0.84 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: 0.83 MATR3matrin 3 (200626_s_at), score: 0.82 MBIPMAP3K12 binding inhibitory protein 1 (218411_s_at), score: 0.84 MEX3Cmex-3 homolog C (C. elegans) (218247_s_at), score: 0.79 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: 0.87 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: 0.83 PCNPPEST proteolytic signal containing nuclear protein (217816_s_at), score: 0.79 RB1CC1RB1-inducible coiled-coil 1 (202034_x_at), score: 0.89 RNF11ring finger protein 11 (208924_at), score: 0.82 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.91 STXBP3syntaxin binding protein 3 (203310_at), score: 0.85 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: 0.86 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.97 TMEM123transmembrane protein 123 (211967_at), score: 0.89 TSNAXtranslin-associated factor X (203983_at), score: 0.87 UBE2D1ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) (211764_s_at), score: 0.88 UFM1ubiquitin-fold modifier 1 (218050_at), score: 0.83 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: 0.81
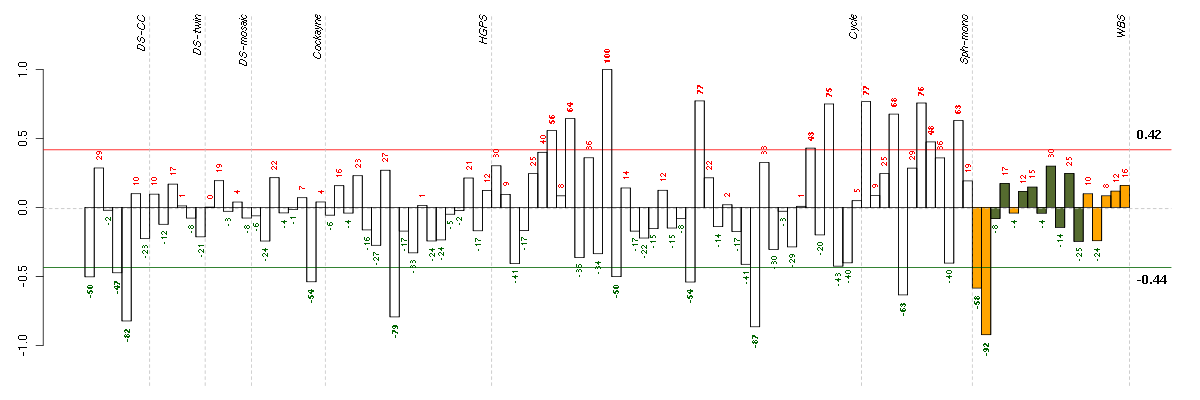
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |