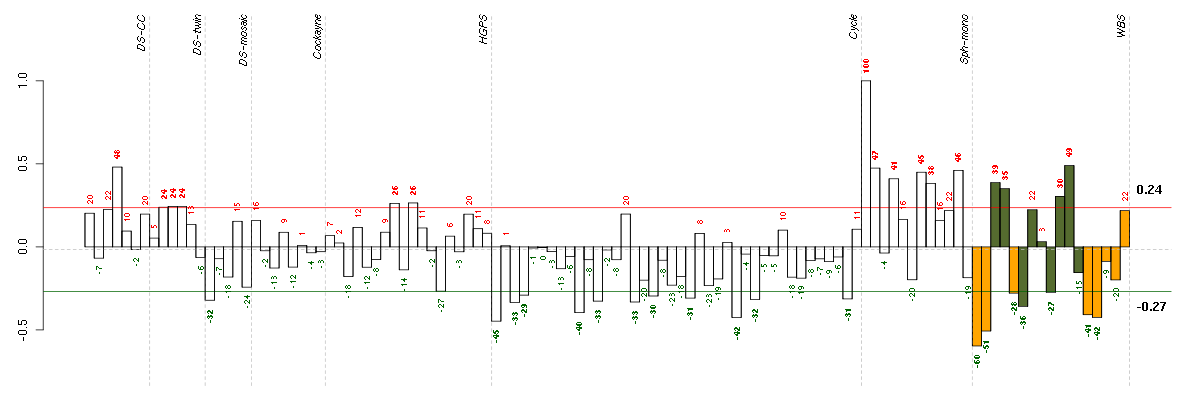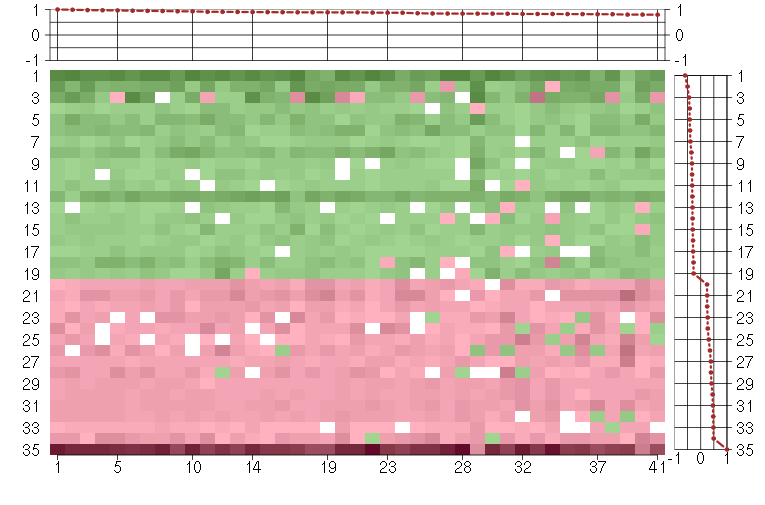

Under-expression is coded with green,
over-expression with red color.



ALMS1Alstrom syndrome 1 (214707_x_at), score: 1 CEP27centrosomal protein 27kDa (220071_x_at), score: 0.89 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: 0.8 CTTNBP2NLCTTNBP2 N-terminal like (214731_at), score: 0.8 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.84 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: 0.99 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (215529_x_at), score: 0.93 FBXW12F-box and WD repeat domain containing 12 (215600_x_at), score: 0.89 FLJ23172hypothetical LOC389177 (217016_x_at), score: 0.89 G3BP1GTPase activating protein (SH3 domain) binding protein 1 (222187_x_at), score: 0.88 HCG2P7HLA complex group 2 pseudogene 7 (216229_x_at), score: 0.94 IBD12Inflammatory bowel disease 12 (215373_x_at), score: 0.98 KIAA0894KIAA0894 protein (207436_x_at), score: 0.91 KLHL24kelch-like 24 (Drosophila) (206551_x_at), score: 0.8 LOC100134401hypothetical protein LOC100134401 (213605_s_at), score: 0.83 LOC152719hypothetical protein LOC152719 (215978_x_at), score: 0.81 LOC23117PI-3-kinase-related kinase SMG-1 isoform 1 homolog (211996_s_at), score: 0.82 LOC647070hypothetical LOC647070 (215467_x_at), score: 0.89 MAP2K7mitogen-activated protein kinase kinase 7 (216206_x_at), score: 0.84 METAP2methionyl aminopeptidase 2 (202015_x_at), score: 0.84 METTL7Amethyltransferase like 7A (211424_x_at), score: 0.93 MFSD11major facilitator superfamily domain containing 11 (221192_x_at), score: 0.91 MKRN1makorin ring finger protein 1 (208082_x_at), score: 0.95 PDE4Cphosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) (206792_x_at), score: 0.89 PGFplacental growth factor (215179_x_at), score: 0.87 POGZpogo transposable element with ZNF domain (215281_x_at), score: 0.95 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (220113_x_at), score: 0.85 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: 0.9 RPL23AP32ribosomal protein L23a pseudogene 32 (207283_at), score: 0.82 RPL35Aribosomal protein L35a (215208_x_at), score: 0.9 SH3BP2SH3-domain binding protein 2 (217257_at), score: 0.83 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: 0.89 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: 0.82 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: 0.84 UBQLN4ubiquilin 4 (222252_x_at), score: 0.96 USP6ubiquitin specific peptidase 6 (Tre-2 oncogene) (206405_x_at), score: 0.82 XRCC2X-ray repair complementing defective repair in Chinese hamster cells 2 (207598_x_at), score: 0.87 ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: 0.97 ZNF160zinc finger protein 160 (214715_x_at), score: 0.9 ZNF492zinc finger protein 492 (215532_x_at), score: 0.84 ZNF611zinc finger protein 611 (208137_x_at), score: 0.83
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| D890_WBS.CEL | 13 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 5042_CNTL.CEL | 6 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 4319_WBS.CEL | 5 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 6089_CNTL.CEL | 9 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 1104_CNTL.CEL | 3 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| 9118_CNTL.CEL | 11 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |