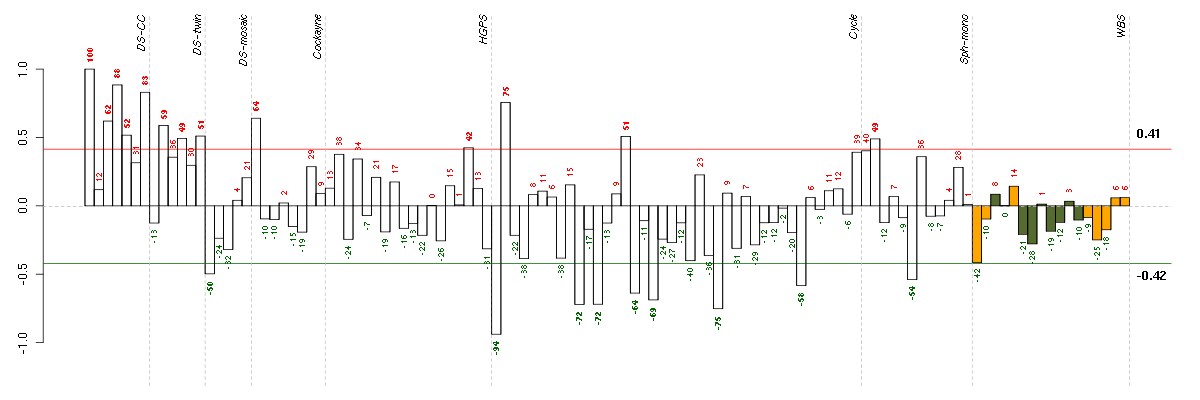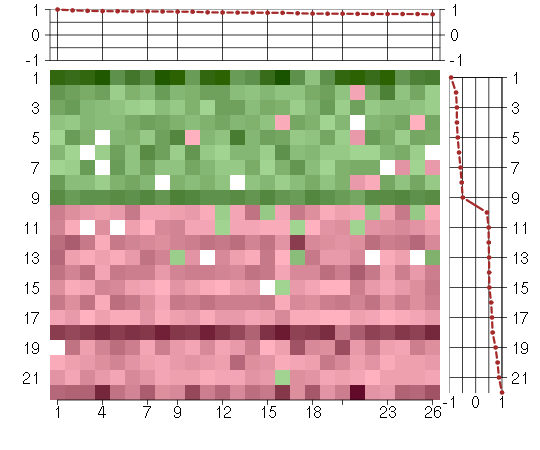

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
sensory perception
The series of events required for an organism to receive a sensory stimulus, convert it to a molecular signal, and recognize and characterize the signal.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
cognition
The operation of the mind by which an organism becomes aware of objects of thought or perception; it includes the mental activities associated with thinking, learning, and memory.
all
This term is the most general term possible


ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: 0.82 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.83 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.91 CA4carbonic anhydrase IV (206209_s_at), score: 0.89 CATSPER2cation channel, sperm associated 2 (217588_at), score: 0.82 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.97 CLDN10claudin 10 (205328_at), score: 0.81 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.93 FAM75A3family with sequence similarity 75, member A3 (215935_at), score: 0.84 HAB1B1 for mucin (215778_x_at), score: 0.84 HSPA6heat shock 70kDa protein 6 (HSP70B') (117_at), score: 0.85 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.88 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: 0.88 MYO1Amyosin IA (211916_s_at), score: 0.87 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.82 OCLMoculomedin (208274_at), score: 0.88 PIPOXpipecolic acid oxidase (221605_s_at), score: 0.93 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: 0.94 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 1 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: 0.93 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 0.95 S100A14S100 calcium binding protein A14 (218677_at), score: 0.87 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: 0.91 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.92 TRIM45tripartite motif-containing 45 (219923_at), score: 0.83 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.83
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |