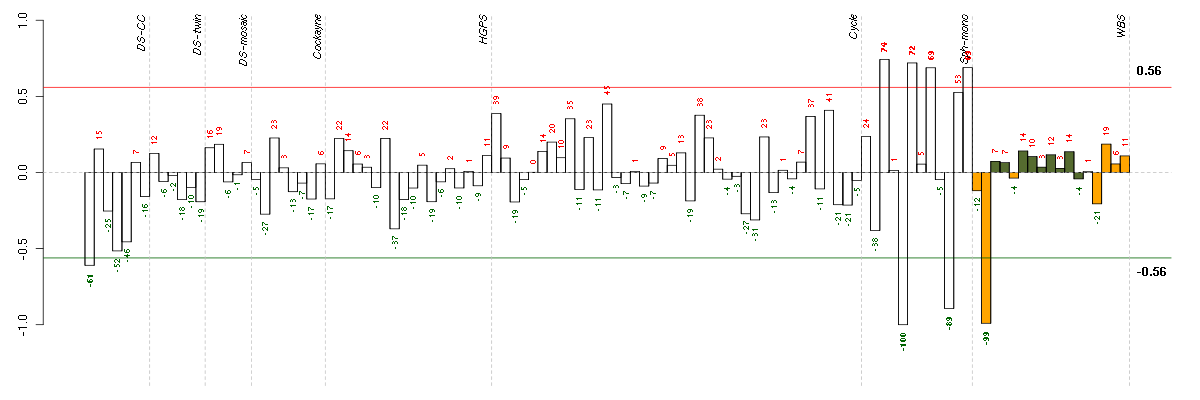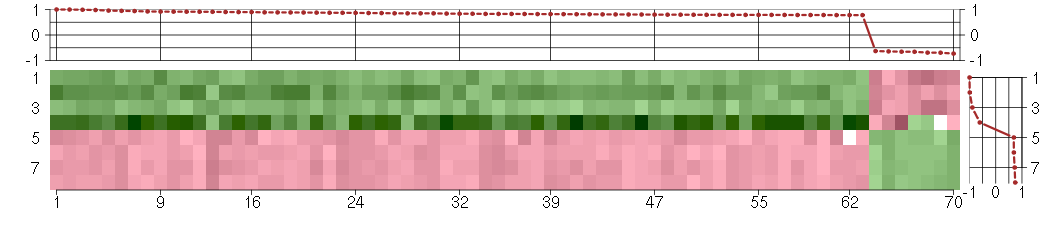

Under-expression is coded with green,
over-expression with red color.



APPBP2amyloid beta precursor protein (cytoplasmic tail) binding protein 2 (202629_at), score: 0.78 ARFGEF2ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) (218098_at), score: 0.84 ARFIP1ADP-ribosylation factor interacting protein 1 (218230_at), score: 0.92 ARHGEF12Rho guanine nucleotide exchange factor (GEF) 12 (201334_s_at), score: 0.8 ARID4AAT rich interactive domain 4A (RBP1-like) (205062_x_at), score: 0.89 ASB9ankyrin repeat and SOCS box-containing 9 (205673_s_at), score: -0.63 ATF1activating transcription factor 1 (222103_at), score: 1 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.79 BAZ2Bbromodomain adjacent to zinc finger domain, 2B (203080_s_at), score: 0.82 BBS10Bardet-Biedl syndrome 10 (219487_at), score: 0.83 C5orf28chromosome 5 open reading frame 28 (219029_at), score: 0.78 C6orf211chromosome 6 open reading frame 211 (218195_at), score: 0.95 CAPZA2capping protein (actin filament) muscle Z-line, alpha 2 (201237_at), score: 0.9 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.73 CEP170centrosomal protein 170kDa (212746_s_at), score: 0.79 CSNK1G3casein kinase 1, gamma 3 (220768_s_at), score: 0.88 CTSDcathepsin D (200766_at), score: -0.66 CUL5cullin 5 (203531_at), score: 0.82 DCUN1D1DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) (218583_s_at), score: 0.9 DPY19L4dpy-19-like 4 (C. elegans) (213391_at), score: 0.81 EML5echinoderm microtubule associated protein like 5 (213063_at), score: 0.8 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.79 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: 0.87 GNG12guanine nucleotide binding protein (G protein), gamma 12 (212294_at), score: 0.79 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 0.91 GRINL1Aglutamate receptor, ionotropic, N-methyl D-aspartate-like 1A (212244_at), score: 0.8 HIF1Ahypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) (200989_at), score: 0.88 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: 0.91 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: 0.83 IMPACTImpact homolog (mouse) (218637_at), score: 0.82 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.79 KCTD9potassium channel tetramerisation domain containing 9 (218823_s_at), score: 0.78 KIAA1128KIAA1128 (209379_s_at), score: 0.81 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: -0.66 MAP4K3mitogen-activated protein kinase kinase kinase kinase 3 (218311_at), score: 0.8 MED23mediator complex subunit 23 (218846_at), score: 0.87 MYO1Bmyosin IB (212364_at), score: 0.87 NRN1neuritin 1 (218625_at), score: -0.64 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: -0.69 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.78 PDS5APDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) (212138_at), score: 0.78 PJA2praja ring finger 2 (201133_s_at), score: 0.92 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (205618_at), score: 0.78 PUM2pumilio homolog 2 (Drosophila) (216221_s_at), score: 0.94 RB1retinoblastoma 1 (203132_at), score: 0.79 RCN2reticulocalbin 2, EF-hand calcium binding domain (201485_s_at), score: 0.83 RIF1RAP1 interacting factor homolog (yeast) (214700_x_at), score: 0.85 RND3Rho family GTPase 3 (212724_at), score: 0.96 ROCK1Rho-associated, coiled-coil containing protein kinase 1 (213044_at), score: 0.86 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: 0.83 SDCCAG1serologically defined colon cancer antigen 1 (218649_x_at), score: 0.85 SENP6SUMO1/sentrin specific peptidase 6 (202318_s_at), score: 0.85 SHOC2soc-2 suppressor of clear homolog (C. elegans) (202777_at), score: 0.89 SLC25A24solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 (204342_at), score: 0.81 SLKSTE20-like kinase (yeast) (206875_s_at), score: 0.91 STAM2signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 (209649_at), score: 0.88 STRN3striatin, calmodulin binding protein 3 (204496_at), score: 1 TCF12transcription factor 12 (208986_at), score: 0.86 TMEM168transmembrane protein 168 (218962_s_at), score: 0.83 TMEM30Atransmembrane protein 30A (217743_s_at), score: 0.98 TMF1TATA element modulatory factor 1 (213024_at), score: 0.92 TOPORStopoisomerase I binding, arginine/serine-rich (204071_s_at), score: 0.79 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: -0.69 USP8ubiquitin specific peptidase 8 (202745_at), score: 0.81 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: 0.82 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (201807_at), score: 0.79 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.8 ZC3H14zinc finger CCCH-type containing 14 (213064_at), score: 0.85 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: 0.9 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: 0.99
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |