

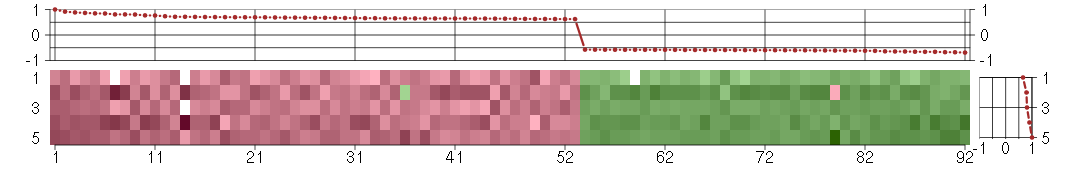

Under-expression is coded with green,
over-expression with red color.

M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
mitotic cell cycle
Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent.
M phase
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the cell cycle comprising nuclear division.
nuclear division
A process by which a cell nucleus is divided into two nuclei, with DNA and other nuclear contents distributed between the daughter nuclei.
organelle organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular component organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a cellular component.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
organelle fission
The creation of two or more organelles by division of one organelle.
cell division
The process resulting in the physical partitioning and separation of a cell into daughter cells.
all
This term is the most general term possible
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromatin
The ordered and organized complex of DNA and protein that forms the chromosome.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
chromosome
A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.

ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: 0.65 AHI1Abelson helper integration site 1 (221569_at), score: 0.67 AIM1absent in melanoma 1 (212543_at), score: -0.59 AREGamphiregulin (205239_at), score: 0.71 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: 0.63 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: -0.58 AURKBaurora kinase B (209464_at), score: -0.6 BMP6bone morphogenetic protein 6 (206176_at), score: 1 C14orf94chromosome 14 open reading frame 94 (218383_at), score: -0.58 C17orf91chromosome 17 open reading frame 91 (214696_at), score: 0.64 C18orf24chromosome 18 open reading frame 24 (217640_x_at), score: -0.59 C2CD2C2 calcium-dependent domain containing 2 (212875_s_at), score: 0.67 CBR3carbonyl reductase 3 (205379_at), score: -0.66 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: 0.62 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: -0.66 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: -0.61 CDCA3cell division cycle associated 3 (221436_s_at), score: -0.59 CENPFcentromere protein F, 350/400ka (mitosin) (207828_s_at), score: -0.62 CEP68centrosomal protein 68kDa (212677_s_at), score: -0.62 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.58 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.67 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: 0.63 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: 0.65 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.71 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: 0.69 ENTPD7ectonucleoside triphosphate diphosphohydrolase 7 (220153_at), score: 0.81 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: 0.7 ETV1ets variant 1 (221911_at), score: -0.59 FAM64Afamily with sequence similarity 64, member A (221591_s_at), score: -0.6 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: 0.87 FGF1fibroblast growth factor 1 (acidic) (205117_at), score: 0.65 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: 0.69 FOXM1forkhead box M1 (202580_x_at), score: -0.58 GHRgrowth hormone receptor (205498_at), score: 0.63 GKglycerol kinase (207387_s_at), score: 0.85 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: 0.92 GPR183G protein-coupled receptor 183 (205419_at), score: 0.81 GPSM2G-protein signaling modulator 2 (AGS3-like, C. elegans) (221922_at), score: -0.61 GRB14growth factor receptor-bound protein 14 (206204_at), score: 0.71 GTSE1G-2 and S-phase expressed 1 (204318_s_at), score: -0.59 H1F0H1 histone family, member 0 (208886_at), score: -0.6 H1FXH1 histone family, member X (204805_s_at), score: -0.65 HJURPHolliday junction recognition protein (218726_at), score: -0.6 HMMRhyaluronan-mediated motility receptor (RHAMM) (207165_at), score: -0.58 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: 0.84 IL33interleukin 33 (209821_at), score: 0.63 IRGQimmunity-related GTPase family, Q (64488_at), score: 0.65 ITGB3BPintegrin beta 3 binding protein (beta3-endonexin) (205176_s_at), score: -0.65 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.7 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: 0.77 KIF15kinesin family member 15 (219306_at), score: -0.6 KIF4Akinesin family member 4A (218355_at), score: -0.61 LIG4ligase IV, DNA, ATP-dependent (206235_at), score: 0.63 MAP2microtubule-associated protein 2 (210015_s_at), score: 0.67 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: 0.7 MFAP3microfibrillar-associated protein 3 (213123_at), score: 0.74 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.62 MKI67antigen identified by monoclonal antibody Ki-67 (212022_s_at), score: -0.57 MLXIPMLX interacting protein (202519_at), score: 0.65 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: 0.65 MNS1meiosis-specific nuclear structural 1 (219703_at), score: -0.58 MREGmelanoregulin (219648_at), score: 0.65 MTMR9myotubularin related protein 9 (204837_at), score: 0.64 NCALDneurocalcin delta (211685_s_at), score: 0.65 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: -0.64 NOX4NADPH oxidase 4 (219773_at), score: 0.65 NUSAP1nucleolar and spindle associated protein 1 (218039_at), score: -0.59 OIP5Opa interacting protein 5 (213599_at), score: -0.58 OTUB2OTU domain, ubiquitin aldehyde binding 2 (219369_s_at), score: 0.7 PCDH9protocadherin 9 (219737_s_at), score: 0.64 PIK3CDphosphoinositide-3-kinase, catalytic, delta polypeptide (203879_at), score: 0.68 POLA1polymerase (DNA directed), alpha 1, catalytic subunit (204835_at), score: -0.58 PTHLHparathyroid hormone-like hormone (211756_at), score: 0.88 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: -0.66 RNASEH2Aribonuclease H2, subunit A (203022_at), score: -0.59 RRP12ribosomal RNA processing 12 homolog (S. cerevisiae) (216913_s_at), score: 0.66 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.68 SLC35A2solute carrier family 35 (UDP-galactose transporter), member A2 (209326_at), score: 0.67 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: 0.7 SMAD3SMAD family member 3 (218284_at), score: -0.67 SNNstannin (218032_at), score: -0.61 TACC3transforming, acidic coiled-coil containing protein 3 (218308_at), score: -0.59 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: -0.68 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: 0.76 TIMELESStimeless homolog (Drosophila) (203046_s_at), score: -0.58 TMEM194Atransmembrane protein 194A (212621_at), score: -0.59 TMEM2transmembrane protein 2 (218113_at), score: 0.68 TOM1target of myb1 (chicken) (202807_s_at), score: 0.66 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: 0.71 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: 0.8 YRDCyrdC domain containing (E. coli) (218647_s_at), score: 0.65 ZNF395zinc finger protein 395 (218149_s_at), score: -0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
