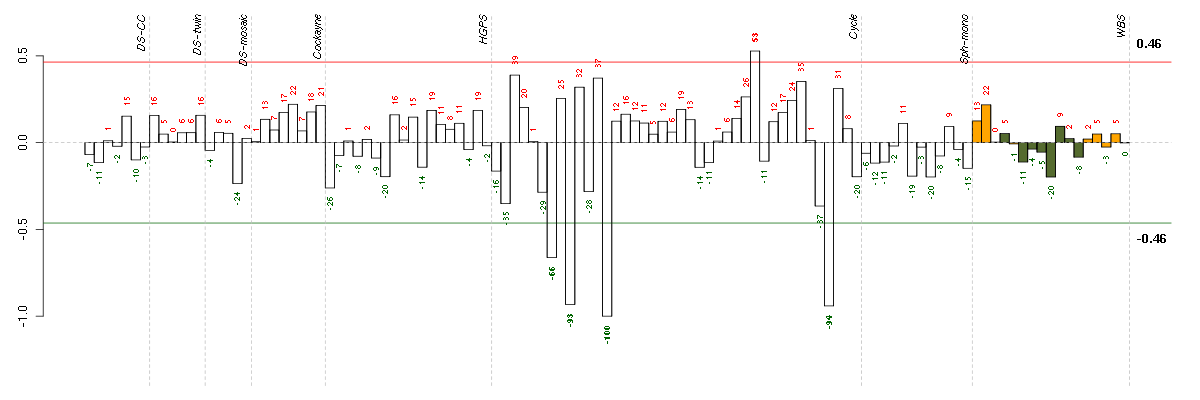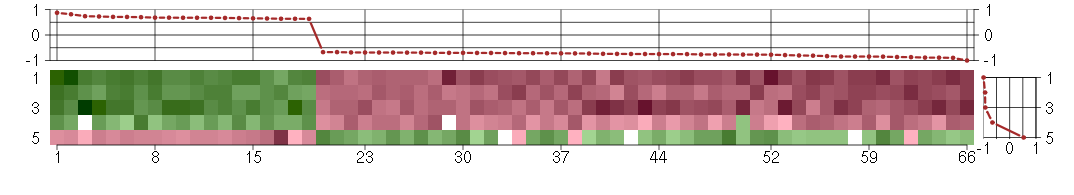

Under-expression is coded with green,
over-expression with red color.



ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.77 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.8 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.65 ADORA2Badenosine A2b receptor (205891_at), score: 0.68 AHI1Abelson helper integration site 1 (221569_at), score: -0.74 AIM1absent in melanoma 1 (212543_at), score: 0.71 ANXA10annexin A10 (210143_at), score: -0.71 BMP6bone morphogenetic protein 6 (206176_at), score: -0.79 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.7 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.85 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.71 CALB2calbindin 2 (205428_s_at), score: -0.68 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.65 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.68 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.87 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.81 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.69 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.89 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.67 EAF2ELL associated factor 2 (219551_at), score: -0.85 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.76 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.71 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.73 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.69 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.68 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.77 GHRgrowth hormone receptor (205498_at), score: -0.87 GKglycerol kinase (207387_s_at), score: -0.75 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.89 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.85 H1FXH1 histone family, member X (204805_s_at), score: 0.68 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.67 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.72 KAL1Kallmann syndrome 1 sequence (205206_at), score: -1 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.72 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.63 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.72 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.83 NCALDneurocalcin delta (211685_s_at), score: -0.75 NEFMneurofilament, medium polypeptide (205113_at), score: -0.86 NOX4NADPH oxidase 4 (219773_at), score: -0.74 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.66 NRXN3neurexin 3 (205795_at), score: -0.7 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.64 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.71 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.69 PCDH9protocadherin 9 (219737_s_at), score: -0.9 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.7 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.76 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.69 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.7 RNF11ring finger protein 11 (208924_at), score: -0.67 SAP30LSAP30-like (219129_s_at), score: 0.71 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.84 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.76 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.75 SMAD3SMAD family member 3 (218284_at), score: 0.73 SNNstannin (218032_at), score: 0.74 SSTR1somatostatin receptor 1 (208482_at), score: -0.74 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.69 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.64 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.75 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.68 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.7 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.77 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.81
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |