
Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
cellular alcohol metabolic process
The chemical reactions and pathways involving alcohols, any of a class of compounds containing one or more hydroxyl groups attached to a saturated carbon atom, as carried out by individual cells.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
steroid biosynthetic process
The chemical reactions and pathways resulting in the formation of steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus; includes de novo formation and steroid interconversion by modification.
cholesterol biosynthetic process
The chemical reactions and pathways resulting in the formation of cholesterol, cholest-5-en-3 beta-ol, the principal sterol of vertebrates and the precursor of many steroids, including bile acids and steroid hormones.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
cholesterol metabolic process
The chemical reactions and pathways involving cholesterol, cholest-5-en-3 beta-ol, the principal sterol of vertebrates and the precursor of many steroids, including bile acids and steroid hormones. It is a component of the plasma membrane lipid bilayer and of plasma lipoproteins and can be found in all animal tissues.
lipid biosynthetic process
The chemical reactions and pathways resulting in the formation of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
sterol metabolic process
The chemical reactions and pathways involving sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
sterol biosynthetic process
The chemical reactions and pathways resulting in the formation of sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
lipid biosynthetic process
The chemical reactions and pathways resulting in the formation of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
steroid biosynthetic process
The chemical reactions and pathways resulting in the formation of steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus; includes de novo formation and steroid interconversion by modification.
sterol metabolic process
The chemical reactions and pathways involving sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
sterol biosynthetic process
The chemical reactions and pathways resulting in the formation of sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
cholesterol biosynthetic process
The chemical reactions and pathways resulting in the formation of cholesterol, cholest-5-en-3 beta-ol, the principal sterol of vertebrates and the precursor of many steroids, including bile acids and steroid hormones.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
cell soma
The portion of a cell bearing surface projections such as axons, dendrites, cilia, or flagella that includes the nucleus, but excludes all cell projections.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

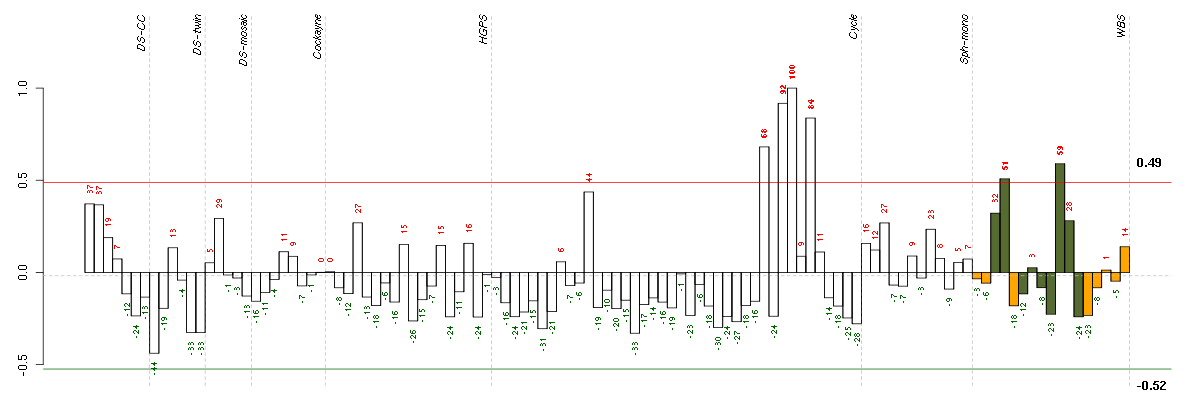
ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (213353_at), score: 0.65 ADAMTS5ADAM metallopeptidase with thrombospondin type 1 motif, 5 (219935_at), score: 0.67 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.86 ANGPTL2angiopoietin-like 2 (213001_at), score: 0.78 ANK2ankyrin 2, neuronal (202920_at), score: 0.79 ARHGAP26Rho GTPase activating protein 26 (205068_s_at), score: 0.64 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.66 B3GALT2UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 (217452_s_at), score: 0.65 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.75 C4orf18chromosome 4 open reading frame 18 (219872_at), score: 0.68 CASP1caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) (211368_s_at), score: 0.69 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: 0.85 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: 0.66 CPA3carboxypeptidase A3 (mast cell) (205624_at), score: 0.75 CPA4carboxypeptidase A4 (205832_at), score: 0.63 CSTF2cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa (204459_at), score: -0.66 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.77 DAPK1death-associated protein kinase 1 (203139_at), score: 0.77 DPYSL2dihydropyrimidinase-like 2 (200762_at), score: 0.63 ECM2extracellular matrix protein 2, female organ and adipocyte specific (206101_at), score: 0.74 ENOX1ecto-NOX disulfide-thiol exchanger 1 (219501_at), score: 0.89 EPHB6EPH receptor B6 (204718_at), score: 0.75 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: 0.68 FRYfurry homolog (Drosophila) (204072_s_at), score: 0.7 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.69 H6PDhexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (221892_at), score: 0.63 HERC6hect domain and RLD 6 (219352_at), score: 0.68 HMGCR3-hydroxy-3-methylglutaryl-Coenzyme A reductase (202540_s_at), score: 0.67 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: 0.7 IDI1isopentenyl-diphosphate delta isomerase 1 (204615_x_at), score: 0.64 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.81 KCNB1potassium voltage-gated channel, Shab-related subfamily, member 1 (211006_s_at), score: 0.77 KCND3potassium voltage-gated channel, Shal-related subfamily, member 3 (213832_at), score: 0.77 LPIN1lipin 1 (212274_at), score: 0.66 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: 0.78 LZTFL1leucine zipper transcription factor-like 1 (218437_s_at), score: 0.7 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.74 MBPmyelin basic protein (210136_at), score: 0.64 MVDmevalonate (diphospho) decarboxylase (203027_s_at), score: 0.71 MX2myxovirus (influenza virus) resistance 2 (mouse) (204994_at), score: 0.78 MYL5myosin, light chain 5, regulatory (205145_s_at), score: 0.66 OLFML2Aolfactomedin-like 2A (213075_at), score: 0.71 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.7 ORC6Lorigin recognition complex, subunit 6 like (yeast) (219105_x_at), score: -0.62 PAPPApregnancy-associated plasma protein A, pappalysin 1 (201981_at), score: 0.67 PDGFDplatelet derived growth factor D (219304_s_at), score: 0.68 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: 0.69 PITPNC1phosphatidylinositol transfer protein, cytoplasmic 1 (219155_at), score: -0.62 PLSCR4phospholipid scramblase 4 (218901_at), score: 0.67 POLR2Dpolymerase (RNA) II (DNA directed) polypeptide D (203664_s_at), score: -0.63 PPLperiplakin (203407_at), score: 1 PRKG1protein kinase, cGMP-dependent, type I (207119_at), score: 0.68 PRPS2phosphoribosyl pyrophosphate synthetase 2 (203401_at), score: -0.63 SEMA3Dsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D (215324_at), score: 0.74 SLC1A3solute carrier family 1 (glial high affinity glutamate transporter), member 3 (202800_at), score: 0.75 SQLEsqualene epoxidase (213562_s_at), score: 0.74 SVEP1sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (213247_at), score: 0.64 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.75 TM4SF1transmembrane 4 L six family member 1 (209386_at), score: -0.62 TM7SF2transmembrane 7 superfamily member 2 (210130_s_at), score: 0.65 TNXAtenascin XA pseudogene (213451_x_at), score: 0.75 TNXBtenascin XB (216333_x_at), score: 0.7 WDR19WD repeat domain 19 (220917_s_at), score: 0.63 WNT2wingless-type MMTV integration site family member 2 (205648_at), score: 0.72 XAF1XIAP associated factor 1 (206133_at), score: 0.68 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: 0.63
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |