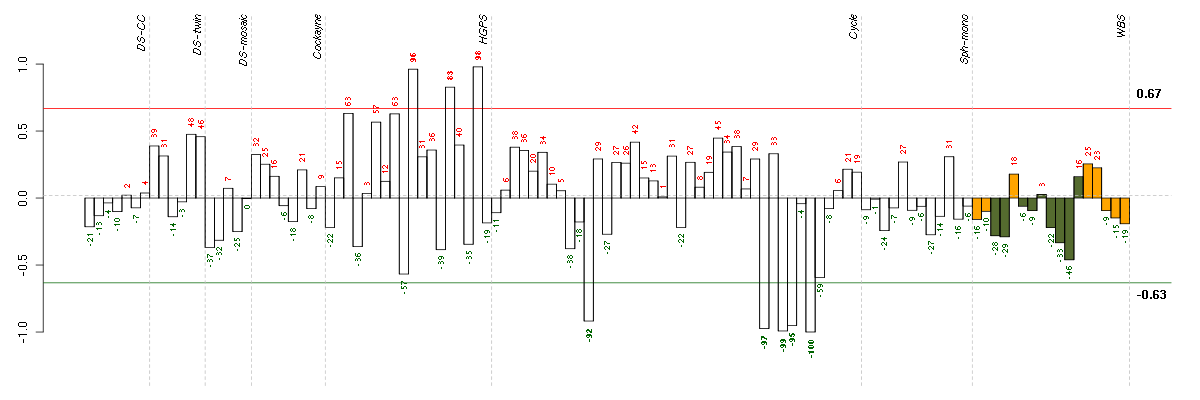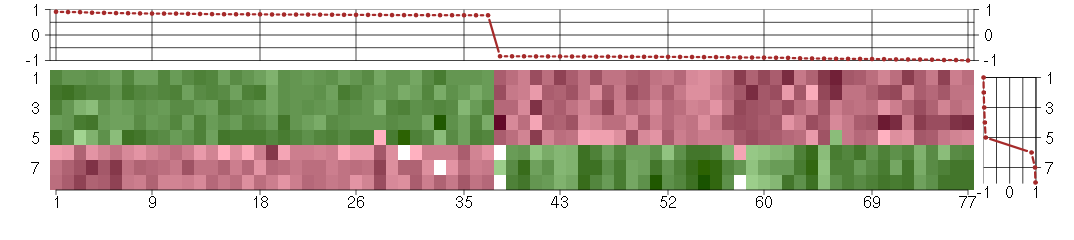

Under-expression is coded with green,
over-expression with red color.

mitotic cell cycle
Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
all
This term is the most general term possible
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosome, centromeric region
The region of a chromosome that includes the centromere and associated proteins. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
chromosome
A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.

ANXA3annexin A3 (209369_at), score: -0.91 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: 0.77 AURKBaurora kinase B (209464_at), score: 0.81 B3GALT2UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 (217452_s_at), score: -0.93 C21orf45chromosome 21 open reading frame 45 (219004_s_at), score: 0.9 C4orf18chromosome 4 open reading frame 18 (219872_at), score: -0.84 CCDC15coiled-coil domain containing 15 (220466_at), score: 0.77 CDC7cell division cycle 7 homolog (S. cerevisiae) (204510_at), score: 0.78 CDCA8cell division cycle associated 8 (221520_s_at), score: 0.78 CDT1chromatin licensing and DNA replication factor 1 (209832_s_at), score: 0.83 CENPEcentromere protein E, 312kDa (205046_at), score: 0.78 CHAF1Bchromatin assembly factor 1, subunit B (p60) (204775_at), score: 0.78 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.8 CORINcorin, serine peptidase (220356_at), score: -0.85 CSTF2cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa (204459_at), score: 0.78 DHCR77-dehydrocholesterol reductase (201790_s_at), score: -0.89 DLEU2Ldeleted in lymphocytic leukemia 2-like (215629_s_at), score: 0.84 DMDdystrophin (203881_s_at), score: -0.94 DRAMdamage-regulated autophagy modulator (218627_at), score: -0.87 DSN1DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) (219512_at), score: 0.91 EXOSC9exosome component 9 (205061_s_at), score: 0.79 EZH2enhancer of zeste homolog 2 (Drosophila) (203358_s_at), score: 0.85 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: -0.88 FRYfurry homolog (Drosophila) (204072_s_at), score: -0.84 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: 0.86 GTSE1G-2 and S-phase expressed 1 (204318_s_at), score: 0.8 HIVEP1human immunodeficiency virus type I enhancer binding protein 1 (204512_at), score: -0.9 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: 0.79 JAG1jagged 1 (Alagille syndrome) (216268_s_at), score: -0.85 KCND3potassium voltage-gated channel, Shal-related subfamily, member 3 (213832_at), score: -0.96 KIF14kinesin family member 14 (206364_at), score: 0.81 KIF18Bkinesin family member 18B (222039_at), score: 0.86 LIG1ligase I, DNA, ATP-dependent (202726_at), score: 0.87 LPHN2latrophilin 2 (206953_s_at), score: -0.87 LPPR4plasticity related gene 1 (213496_at), score: -0.84 LRRC17leucine rich repeat containing 17 (205381_at), score: -0.84 MBPmyelin basic protein (210136_at), score: -0.95 MCM10minichromosome maintenance complex component 10 (220651_s_at), score: 0.81 MCM4minichromosome maintenance complex component 4 (212141_at), score: 0.8 MGPmatrix Gla protein (202291_s_at), score: -0.87 MKI67antigen identified by monoclonal antibody Ki-67 (212022_s_at), score: 0.79 NRIP3nuclear receptor interacting protein 3 (219557_s_at), score: 0.9 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: -0.96 ORC6Lorigin recognition complex, subunit 6 like (yeast) (219105_x_at), score: 0.8 PDCD4programmed cell death 4 (neoplastic transformation inhibitor) (202731_at), score: -0.85 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: -0.97 PGRMC2progesterone receptor membrane component 2 (213227_at), score: -0.86 PIRpirin (iron-binding nuclear protein) (207469_s_at), score: -0.84 PLA2G16phospholipase A2, group XVI (209581_at), score: -0.86 PLSCR4phospholipid scramblase 4 (218901_at), score: -0.94 PNMA2paraneoplastic antigen MA2 (209598_at), score: -0.85 POLA1polymerase (DNA directed), alpha 1, catalytic subunit (204835_at), score: 0.79 PPLperiplakin (203407_at), score: -0.83 PRKG1protein kinase, cGMP-dependent, type I (207119_at), score: -0.93 PSMC3IPPSMC3 interacting protein (213951_s_at), score: 0.79 PTGER4prostaglandin E receptor 4 (subtype EP4) (204897_at), score: -0.85 RAD54BRAD54 homolog B (S. cerevisiae) (219494_at), score: 0.83 RBL1retinoblastoma-like 1 (p107) (205296_at), score: 0.84 RFWD3ring finger and WD repeat domain 3 (218564_at), score: 0.87 ROR1receptor tyrosine kinase-like orphan receptor 1 (205805_s_at), score: -0.86 RRAGBRas-related GTP binding B (205540_s_at), score: -0.86 SC4MOLsterol-C4-methyl oxidase-like (209146_at), score: -0.89 SCARA3scavenger receptor class A, member 3 (219416_at), score: 0.78 SQLEsqualene epoxidase (213562_s_at), score: -0.88 SSX2IPsynovial sarcoma, X breakpoint 2 interacting protein (203016_s_at), score: 0.8 STK38Lserine/threonine kinase 38 like (212572_at), score: -0.84 THBS2thrombospondin 2 (203083_at), score: -0.86 TMEM194Atransmembrane protein 194A (212621_at), score: 0.82 TNXAtenascin XA pseudogene (213451_x_at), score: -1 TNXBtenascin XB (216333_x_at), score: -0.98 TSC22D3TSC22 domain family, member 3 (208763_s_at), score: -0.98 TTKTTK protein kinase (204822_at), score: 0.78 TUFT1tuftelin 1 (205807_s_at), score: -0.96 WHSC1Wolf-Hirschhorn syndrome candidate 1 (209053_s_at), score: 0.84 WRAP53WD repeat containing, antisense to TP53 (44563_at), score: 0.82 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: -0.94 ZNF423zinc finger protein 423 (214761_at), score: -0.92
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |