

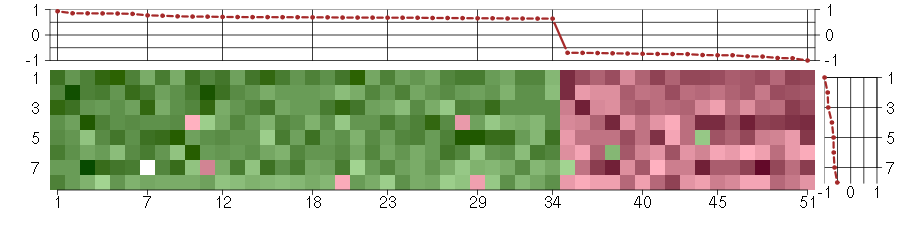

Under-expression is coded with green,
over-expression with red color.

RNA localization
A process by which RNA is transported to, or maintained in, a specific location.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
intracellular mRNA localization
Any process by which mRNA is transported to, or maintained in, a specific location within the cell.
macromolecule localization
Any process by which a macromolecule is transported to, or maintained in, a specific location.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
all
This term is the most general term possible


ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.65 ADCY3adenylate cyclase 3 (209321_s_at), score: 0.72 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.65 BIN1bridging integrator 1 (210201_x_at), score: 0.71 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.68 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.68 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.68 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.85 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.66 EPHA4EPH receptor A4 (206114_at), score: -0.75 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.7 FOXK2forkhead box K2 (203064_s_at), score: 0.93 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.79 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: 0.72 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.9 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.84 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.72 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: -0.74 KIAA1305KIAA1305 (220911_s_at), score: 0.69 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.79 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.7 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.79 MC4Rmelanocortin 4 receptor (221467_at), score: -0.91 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.64 NEFMneurofilament, medium polypeptide (205113_at), score: -1 NUDT11nudix (nucleoside diphosphate linked moiety X)-type motif 11 (219855_at), score: 0.68 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.84 PCDH9protocadherin 9 (219737_s_at), score: -0.85 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: 0.66 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.68 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.67 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.64 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.85 RNF220ring finger protein 220 (219988_s_at), score: 0.7 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.74 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.76 SF1splicing factor 1 (208313_s_at), score: 0.7 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.77 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.66 SNX27sorting nexin family member 27 (221006_s_at), score: 0.7 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (207320_x_at), score: 0.67 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.71 TMEM149transmembrane protein 149 (219690_at), score: -0.75 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: 0.68 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: -0.7 TRIM21tripartite motif-containing 21 (204804_at), score: 0.73 TSKUtsukushin (218245_at), score: 0.85 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.69 WDR6WD repeat domain 6 (217734_s_at), score: 0.83 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.7 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.73
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
