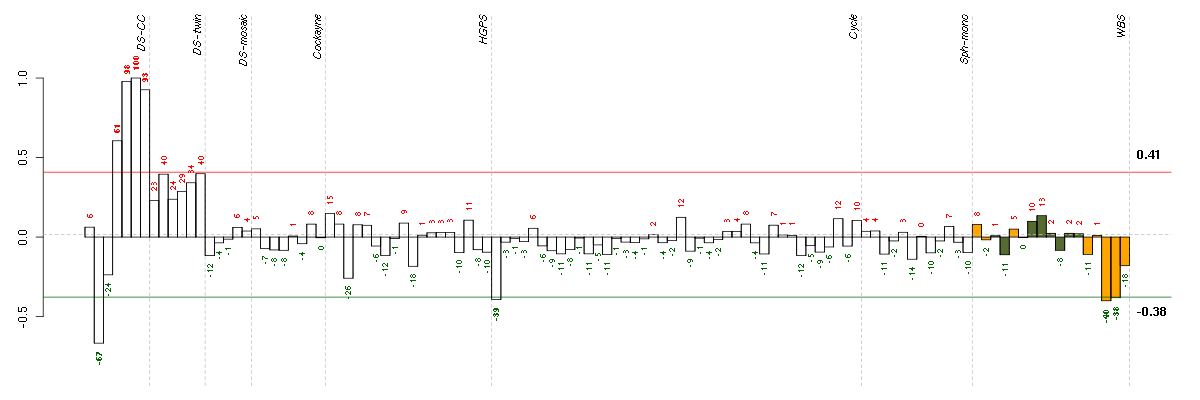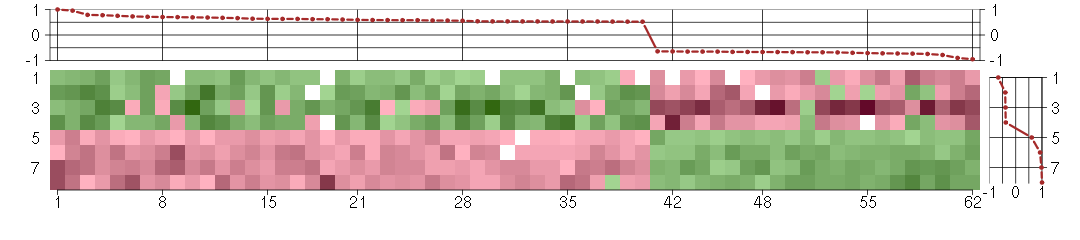

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

AIPL1aryl hydrocarbon receptor interacting protein-like 1 (219977_at), score: 0.54 AK3L1adenylate kinase 3-like 1 (204348_s_at), score: 0.56 ANKRD11ankyrin repeat domain 11 (219437_s_at), score: 0.52 APOMapolipoprotein M (205682_x_at), score: 0.53 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: 0.53 ASAP2ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 (206414_s_at), score: -0.78 B4GALT6UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (206233_at), score: 0.67 BNIP3BCL2/adenovirus E1B 19kDa interacting protein 3 (201848_s_at), score: 0.63 C1orf115chromosome 1 open reading frame 115 (218546_at), score: 0.79 CAB39calcium binding protein 39 (217873_at), score: -0.65 CCND2cyclin D2 (200953_s_at), score: 0.95 CCRL1chemokine (C-C motif) receptor-like 1 (220351_at), score: -0.9 CD302CD302 molecule (203799_at), score: -0.64 CDH18cadherin 18, type 2 (206280_at), score: 0.7 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (203341_at), score: -0.67 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: -0.74 CPNE3copine III (202118_s_at), score: -0.65 CTSL1cathepsin L1 (202087_s_at), score: -0.67 CYBBcytochrome b-245, beta polypeptide (203923_s_at), score: 0.69 EDARectodysplasin A receptor (220048_at), score: 0.53 EIF5Aeukaryotic translation initiation factor 5A (201123_s_at), score: 0.78 ENO2enolase 2 (gamma, neuronal) (201313_at), score: 0.59 ETNK2ethanolamine kinase 2 (219268_at), score: 0.53 F10coagulation factor X (205620_at), score: -0.64 FAM38Bfamily with sequence similarity 38, member B (219602_s_at), score: 0.63 FLGfilaggrin (215704_at), score: 1 FNDC3Afibronectin type III domain containing 3A (202304_at), score: -0.67 FOSv-fos FBJ murine osteosarcoma viral oncogene homolog (209189_at), score: 0.73 GOLSYNGolgi-localized protein (218692_at), score: 0.53 HOXC8homeobox C8 (221350_at), score: 0.53 HPCAL1hippocalcin-like 1 (205462_s_at), score: -0.68 IFITM1interferon induced transmembrane protein 1 (9-27) (214022_s_at), score: -0.68 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: -0.73 JAM2junctional adhesion molecule 2 (219213_at), score: 0.7 LHFPlipoma HMGIC fusion partner (218656_s_at), score: -0.95 LOXL2lysyl oxidase-like 2 (202997_s_at), score: 0.63 LPHN2latrophilin 2 (206953_s_at), score: 0.53 LY96lymphocyte antigen 96 (206584_at), score: -0.67 MEOX1mesenchyme homeobox 1 (205619_s_at), score: 0.58 MESTmesoderm specific transcript homolog (mouse) (202016_at), score: 0.58 MUC3Amucin 3A, cell surface associated (217117_x_at), score: 0.59 NLGN4Xneuroligin 4, X-linked (221933_at), score: 0.75 NT5DC25'-nucleotidase domain containing 2 (218051_s_at), score: 0.53 OCLMoculomedin (208274_at), score: 0.57 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (207543_s_at), score: 0.66 PALMparalemmin (203859_s_at), score: 0.57 PAPPA2pappalysin 2 (213332_at), score: 0.69 PFKLphosphofructokinase, liver (201102_s_at), score: 0.71 PNRC2proline-rich nuclear receptor coactivator 2 (217779_s_at), score: -0.7 POSTNperiostin, osteoblast specific factor (210809_s_at), score: 0.64 PPARDperoxisome proliferator-activated receptor delta (37152_at), score: -0.71 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: 0.52 RCAN2regulator of calcineurin 2 (203498_at), score: -0.73 RELBv-rel reticuloendotheliosis viral oncogene homolog B (205205_at), score: -0.65 SCRG1scrapie responsive protein 1 (205475_at), score: 0.62 SFRP4secreted frizzled-related protein 4 (204051_s_at), score: 0.62 SOD3superoxide dismutase 3, extracellular (205236_x_at), score: -0.72 SOX11SRY (sex determining region Y)-box 11 (204914_s_at), score: 0.53 TNFRSF1Btumor necrosis factor receptor superfamily, member 1B (203508_at), score: -0.66 TPI1triosephosphate isomerase 1 (213011_s_at), score: 0.61 UCHL1ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (201387_s_at), score: 0.58 WISP2WNT1 inducible signaling pathway protein 2 (205792_at), score: -0.68
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |