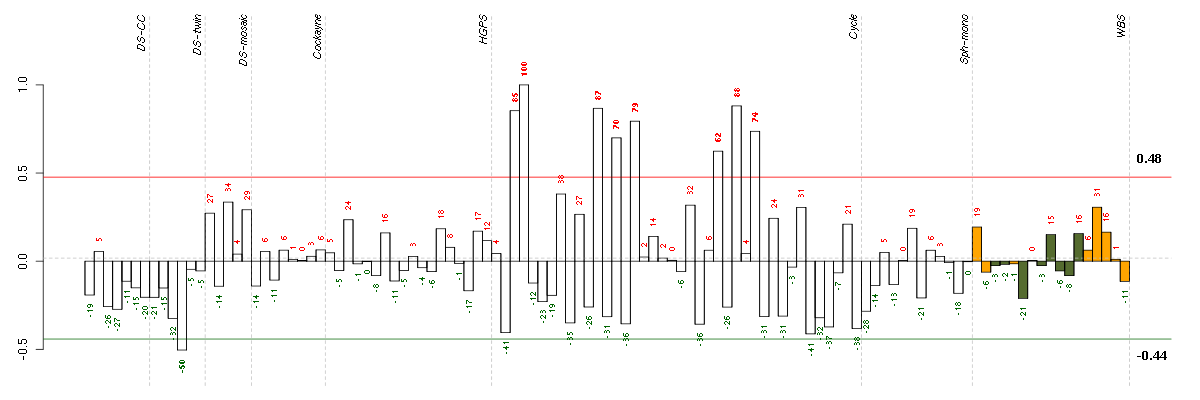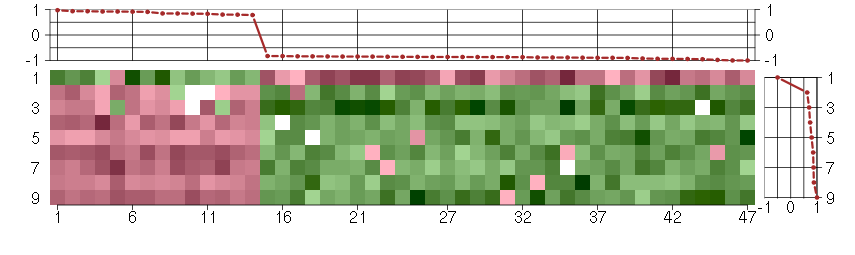

Under-expression is coded with green,
over-expression with red color.



ACTN3actinin, alpha 3 (206891_at), score: -0.87 APOC1apolipoprotein C-I (213553_x_at), score: -0.85 BCL2L1BCL2-like 1 (215037_s_at), score: 0.8 COL16A1collagen, type XVI, alpha 1 (204345_at), score: -0.85 CPNE3copine III (202118_s_at), score: 0.91 DCBLD2discoidin, CUB and LCCL domain containing 2 (213865_at), score: 0.78 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.86 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: -0.83 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: -0.86 FER1L4fer-1-like 4 (C. elegans) (222245_s_at), score: -0.84 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: -0.86 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: -1 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: -0.88 GPR144G protein-coupled receptor 144 (216289_at), score: -1 HAB1B1 for mucin (215778_x_at), score: -0.94 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.85 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: -0.98 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.93 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: -0.88 IL9Rinterleukin 9 receptor (214950_at), score: -0.89 KIF26Bkinesin family member 26B (220002_at), score: -0.9 KLF7Kruppel-like factor 7 (ubiquitous) (204334_at), score: -0.89 LAT2linker for activation of T cells family, member 2 (211768_at), score: -0.86 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: -0.86 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.83 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.92 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.89 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.84 PARP6poly (ADP-ribose) polymerase family, member 6 (219639_x_at), score: -0.84 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.84 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (216347_s_at), score: -0.87 PRRX1paired related homeobox 1 (205991_s_at), score: 0.84 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.88 PXNpaxillin (211823_s_at), score: 0.97 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: 0.92 SAA4serum amyloid A4, constitutive (207096_at), score: -0.93 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.8 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.85 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: 0.93 SLC2A3solute carrier family 2 (facilitated glucose transporter), member 3 (202499_s_at), score: -0.94 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: -0.95 SSX3synovial sarcoma, X breakpoint 3 (211731_x_at), score: -0.92 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: -0.85 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.91 TRAPPC6Atrafficking protein particle complex 6A (204985_s_at), score: -0.87 ZNF652zinc finger protein 652 (205594_at), score: -0.84 ZNF783zinc finger family member 783 (221876_at), score: -0.83
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |