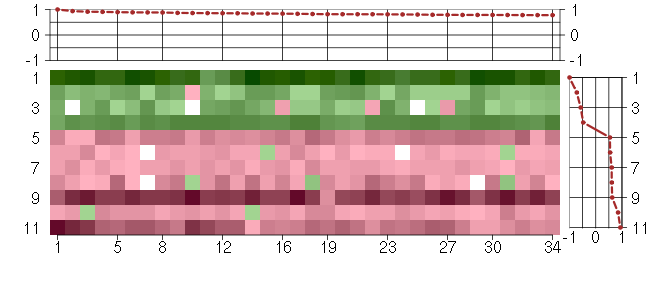

Under-expression is coded with green,
over-expression with red color.



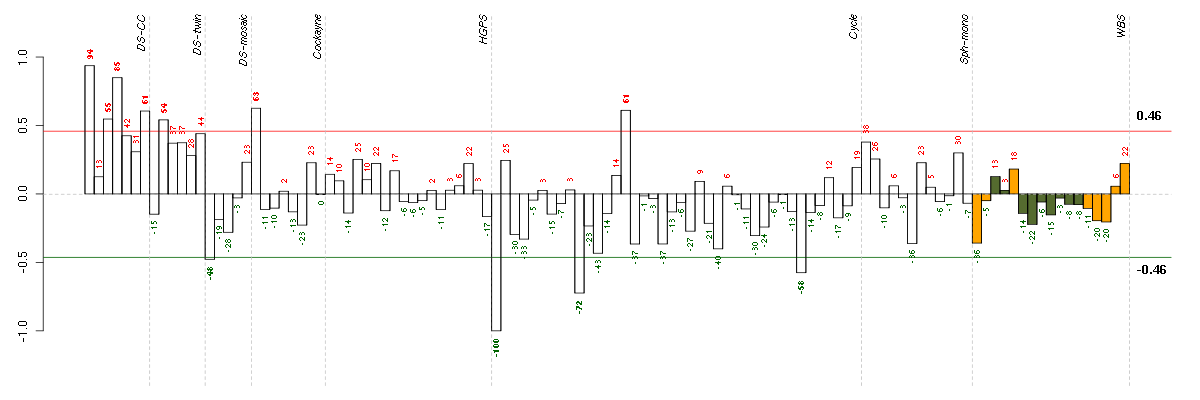
C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.86 C8orf60chromosome 8 open reading frame 60 (220712_at), score: 0.81 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.81 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: 0.78 CHRNA6cholinergic receptor, nicotinic, alpha 6 (207568_at), score: 0.84 CYBBcytochrome b-245, beta polypeptide (203923_s_at), score: 0.84 DHRS9dehydrogenase/reductase (SDR family) member 9 (219799_s_at), score: 0.79 GPATCH4G patch domain containing 4 (220596_at), score: 0.81 KCNQ3potassium voltage-gated channel, KQT-like subfamily, member 3 (206573_at), score: 0.82 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: 0.88 LOC643293FLJ44451 pseudogene (215057_at), score: 0.79 MBmyoglobin (204179_at), score: 0.78 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: 0.84 MMRN2multimerin 2 (219091_s_at), score: 0.86 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.89 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: 0.81 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.81 MYO1Amyosin IA (211916_s_at), score: 0.91 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: 0.79 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: 0.79 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.82 PIPOXpipecolic acid oxidase (221605_s_at), score: 0.85 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: 0.94 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.9 RETret proto-oncogene (205879_x_at), score: 0.78 RP11-35N6.1plasticity related gene 3 (219732_at), score: 0.83 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 0.91 S100A14S100 calcium binding protein A14 (218677_at), score: 0.87 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: 0.8 TACR1tachykinin receptor 1 (208048_at), score: 0.89 TBX6T-box 6 (207684_at), score: 0.85 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.78 TP63tumor protein p63 (209863_s_at), score: 0.79 VGFVGF nerve growth factor inducible (205586_x_at), score: 1
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |