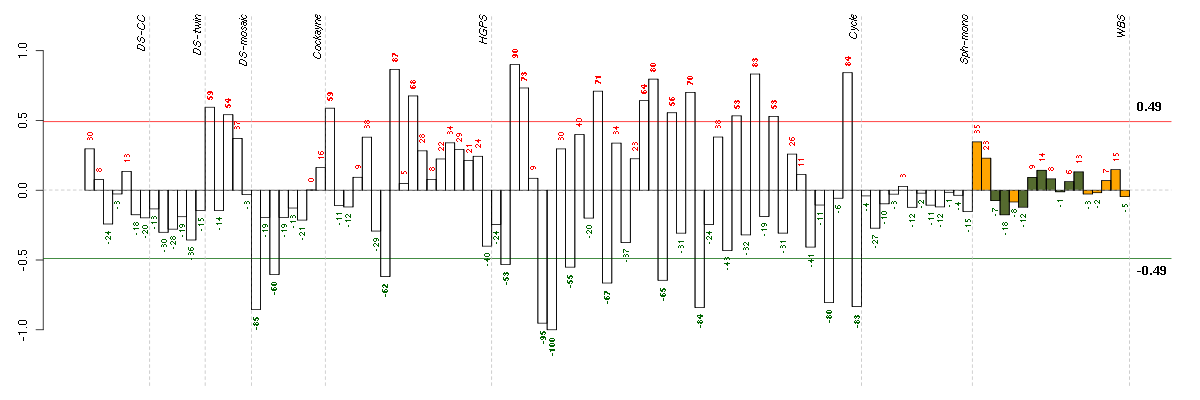

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
transcription
The synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The synthesis of RNA on a template of DNA.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of DNA-dependent transcription.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
response to nutrient
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to extracellular stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an extracellular stimulus.
gene expression
The process by which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
RNA metabolic process
The chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
response to nutrient levels
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus reflecting the presence, absence, or concentration of nutrients.
cellular response to extracellular stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an extracellular stimulus.
cellular response to nutrient levels
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus reflecting the presence, absence, or concentration of nutrients.
cellular response to nutrient
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
response to chemical stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins.
biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription by carbon catabolites
Any process involving carbon catabolites that modulates the frequency, rate or extent of transcription.
regulation of transcription by glucose
Any process involving glucose that modulates the frequency, rate or extent or transcription.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular response to extracellular stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an extracellular stimulus.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
transcription
The synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
cellular response to extracellular stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an extracellular stimulus.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The synthesis of RNA on a template of DNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
RNA metabolic process
The chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
cellular response to nutrient
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
response to nutrient
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
cellular response to nutrient levels
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus reflecting the presence, absence, or concentration of nutrients.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of DNA-dependent transcription.
transcription
The synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of DNA-dependent transcription.
regulation of transcription by carbon catabolites
Any process involving carbon catabolites that modulates the frequency, rate or extent of transcription.


ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.68 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.64 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.66 ARSAarylsulfatase A (204443_at), score: 0.77 ATN1atrophin 1 (40489_at), score: 0.8 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.78 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.65 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.8 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.68 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.65 CABIN1calcineurin binding protein 1 (37652_at), score: 0.67 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.89 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.74 CENPTcentromere protein T (218148_at), score: 0.65 CHMP1Achromatin modifying protein 1A (201933_at), score: 0.63 CICcapicua homolog (Drosophila) (212784_at), score: 0.74 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.67 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.71 DNM2dynamin 2 (202253_s_at), score: 0.66 DOK4docking protein 4 (209691_s_at), score: 0.71 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.66 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.8 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.67 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.65 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.66 FOXK2forkhead box K2 (203064_s_at), score: 0.84 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.81 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.71 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.74 HSF1heat shock transcription factor 1 (202344_at), score: 0.63 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.76 JUNDjun D proto-oncogene (203751_x_at), score: 0.66 JUPjunction plakoglobin (201015_s_at), score: 0.68 LEPREL2leprecan-like 2 (204854_at), score: 0.71 LMF2lipase maturation factor 2 (212682_s_at), score: 0.66 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.66 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.99 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.69 MAP4microtubule-associated protein 4 (200836_s_at), score: 0.67 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.78 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.69 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.67 NCDNneurochondrin (209556_at), score: 0.67 NCKAP1NCK-associated protein 1 (217465_at), score: -0.87 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: 0.63 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.64 PARVBparvin, beta (204629_at), score: 0.8 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.79 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.8 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 1 PKN1protein kinase N1 (202161_at), score: 0.69 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.69 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.69 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.75 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.71 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.64 RBM15BRNA binding motif protein 15B (202689_at), score: 0.63 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.83 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.82 SBF1SET binding factor 1 (39835_at), score: 0.64 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.68 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.98 SF1splicing factor 1 (208313_s_at), score: 0.7 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.75 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.64 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.78 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.71 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.76 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.74 SUPT6Hsuppressor of Ty 6 homolog (S. cerevisiae) (208831_x_at), score: 0.66 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.83 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.64 TSKUtsukushin (218245_at), score: 0.67 TXLNAtaxilin alpha (212300_at), score: 0.77 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.71 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.69 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.93 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.82 WDR42AWD repeat domain 42A (202249_s_at), score: 0.67 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.65 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.64
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |