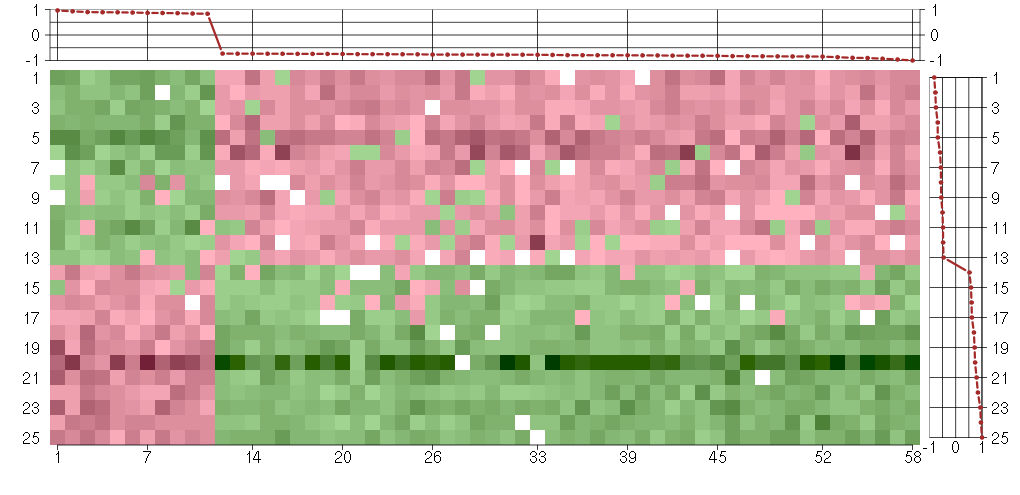

Under-expression is coded with green,
over-expression with red color.



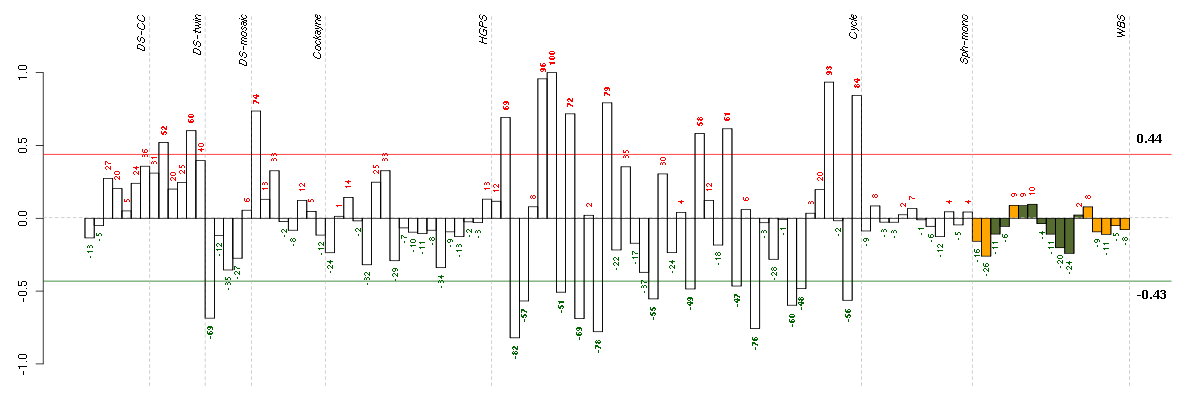
ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: -0.82 AGRNagrin (217419_x_at), score: -0.77 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.87 ARSAarylsulfatase A (204443_at), score: -0.77 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.83 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: -0.81 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.76 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: -0.79 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: -0.77 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.83 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: -0.84 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: 0.83 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.76 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: -0.75 FOXF1forkhead box F1 (205935_at), score: -0.73 FOXK2forkhead box K2 (203064_s_at), score: -0.93 GMPRguanosine monophosphate reductase (204187_at), score: -0.77 HAB1B1 for mucin (215778_x_at), score: 0.89 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.89 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.83 JUNDjun D proto-oncogene (203751_x_at), score: -0.79 JUPjunction plakoglobin (201015_s_at), score: -0.9 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.96 KIAA1305KIAA1305 (220911_s_at), score: -0.89 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: 0.86 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: -0.77 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.84 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.86 NEFMneurofilament, medium polypeptide (205113_at), score: 0.9 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: -0.73 NINJ1ninjurin 1 (203045_at), score: -0.78 PCDHG@protocadherin gamma cluster (215836_s_at), score: -0.75 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.77 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.87 PKN1protein kinase N1 (202161_at), score: -0.79 PLAUplasminogen activator, urokinase (211668_s_at), score: -0.74 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: -0.8 PXNpaxillin (211823_s_at), score: -0.75 RAB11BRAB11B, member RAS oncogene family (34478_at), score: -0.73 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.84 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.88 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.8 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -1 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: -0.74 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: -0.77 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.78 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.8 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: -0.74 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.74 TSC2tuberous sclerosis 2 (215735_s_at), score: -0.74 TSKUtsukushin (218245_at), score: -0.85 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: 0.93 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.81 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.96 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.8 WBP2WW domain binding protein 2 (209117_at), score: -0.73 WDR42AWD repeat domain 42A (202249_s_at), score: -0.81 WDR6WD repeat domain 6 (217734_s_at), score: -0.84
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |