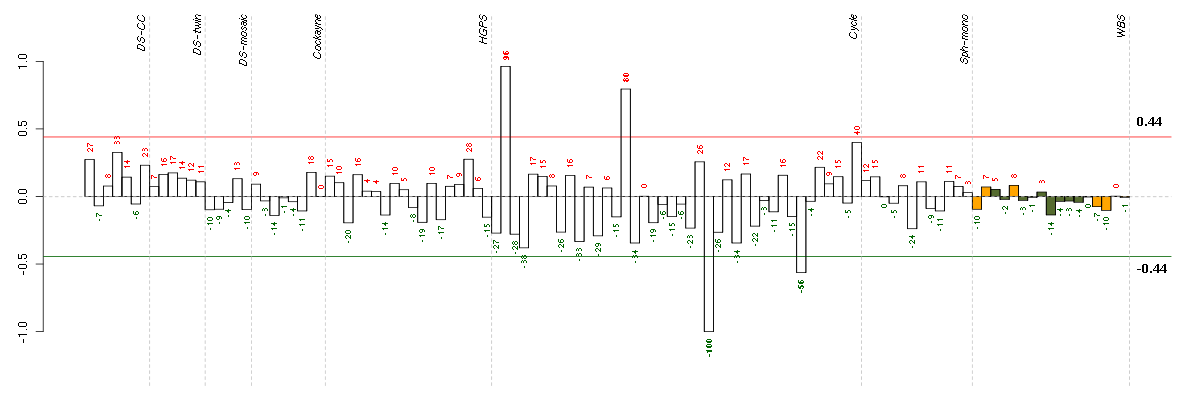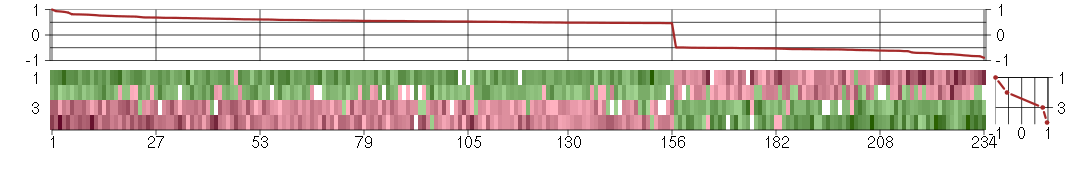

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
receptor binding
Interacting selectively with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
hormone activity
The action characteristic of a hormone, any substance formed in very small amounts in one specialized organ or group of cells and carried (sometimes in the bloodstream) to another organ or group of cells in the same organism, upon which it has a specific regulatory action. The term was originally applied to agents with a stimulatory physiological action in vertebrate animals (as opposed to a chalone, which has a depressant action). Usage is now extended to regulatory compounds in lower animals and plants, and to synthetic substances having comparable effects; all bind receptors and trigger some biological process.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.92 ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: -0.55 ACTR8ARP8 actin-related protein 8 homolog (yeast) (218658_s_at), score: -0.52 ADAM21ADAM metallopeptidase domain 21 (207665_at), score: -0.53 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.69 ADORA1adenosine A1 receptor (216220_s_at), score: 0.69 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: 0.62 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.5 ALS2CR8amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 8 (219834_at), score: -0.56 ANKRD46ankyrin repeat domain 46 (212731_at), score: -0.59 APOC4apolipoprotein C-IV (206738_at), score: 0.65 APPamyloid beta (A4) precursor protein (214953_s_at), score: 0.94 ARID3AAT rich interactive domain 3A (BRIGHT-like) (205865_at), score: -0.49 ARL15ADP-ribosylation factor-like 15 (219842_at), score: -0.54 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.64 BAI2brain-specific angiogenesis inhibitor 2 (204966_at), score: 0.54 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.69 BCHEbutyrylcholinesterase (205433_at), score: -0.61 BCL2B-cell CLL/lymphoma 2 (203685_at), score: -0.53 BEND5BEN domain containing 5 (219670_at), score: 0.48 BEST1bestrophin 1 (207671_s_at), score: 0.59 BNIP1BCL2/adenovirus E1B 19kDa interacting protein 1 (204930_s_at), score: -0.49 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: -0.83 BTF3L2basic transcription factor 3, like 2 (217461_x_at), score: 0.48 C18orf1chromosome 18 open reading frame 1 (209574_s_at), score: -0.7 C1orf69chromosome 1 open reading frame 69 (215490_at), score: 0.55 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: 0.53 C4Acomplement component 4A (Rodgers blood group) (214428_x_at), score: 0.63 CACNA1Gcalcium channel, voltage-dependent, T type, alpha 1G subunit (211315_s_at), score: 0.68 CASP8caspase 8, apoptosis-related cysteine peptidase (213373_s_at), score: -0.54 CATSPER2cation channel, sperm associated 2 (217588_at), score: 0.56 CC2D1Acoiled-coil and C2 domain containing 1A (58994_at), score: 0.67 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.73 CCDC68coiled-coil domain containing 68 (220180_at), score: -0.55 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.58 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.62 CDO1cysteine dioxygenase, type I (204154_at), score: -0.51 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: 0.55 CLCN2chloride channel 2 (213499_at), score: 0.54 CNNM3cyclin M3 (220739_s_at), score: -0.62 CNTLNcentlein, centrosomal protein (220095_at), score: -0.61 CROCCciliary rootlet coiled-coil, rootletin (206274_s_at), score: 0.51 CTAGE1cutaneous T-cell lymphoma-associated antigen 1 (220957_at), score: -0.55 CTSZcathepsin Z (210042_s_at), score: 0.49 CYorf15Bchromosome Y open reading frame 15B (214131_at), score: 0.71 CYTH4cytohesin 4 (219183_s_at), score: 0.57 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: 0.48 DEM1defects in morphology 1 homolog (S. cerevisiae) (219567_s_at), score: 0.47 DHX34DEAH (Asp-Glu-Ala-His) box polypeptide 34 (214017_s_at), score: 0.55 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.54 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.56 EDAectodysplasin A (211130_x_at), score: 0.73 EDNRBendothelin receptor type B (204271_s_at), score: -0.51 EFNB1ephrin-B1 (202711_at), score: 0.54 ELAC1elaC homolog 1 (E. coli) (219325_s_at), score: 0.49 EPHA3EPH receptor A3 (206070_s_at), score: -0.52 EPHB4EPH receptor B4 (216680_s_at), score: 0.56 EPN1epsin 1 (221141_x_at), score: 0.65 EPOerythropoietin (217254_s_at), score: 0.59 EPS8L1EPS8-like 1 (218778_x_at), score: 0.72 EXOC2exocyst complex component 2 (219349_s_at), score: -0.51 EXPH5exophilin 5 (214734_at), score: -0.71 FAM59Afamily with sequence similarity 59, member A (219377_at), score: -0.52 FBXO2F-box protein 2 (219305_x_at), score: 0.48 FGF13fibroblast growth factor 13 (205110_s_at), score: -0.56 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 1 FUSfusion (involved in t(12;16) in malignant liposarcoma) (217370_x_at), score: 0.49 FXYD2FXYD domain containing ion transport regulator 2 (207434_s_at), score: 0.48 GABRR2gamma-aminobutyric acid (GABA) receptor, rho 2 (208217_at), score: 0.48 GARNL1GTPase activating Rap/RanGAP domain-like 1 (214855_s_at), score: -0.59 GASTgastrin (208138_at), score: 0.81 GATMglycine amidinotransferase (L-arginine:glycine amidinotransferase) (203178_at), score: -0.62 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.57 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: 0.55 GGTLC1gamma-glutamyltransferase light chain 1 (211416_x_at), score: 0.51 GLRA1glycine receptor, alpha 1 (207972_at), score: 0.47 GNB1Lguanine nucleotide binding protein (G protein), beta polypeptide 1-like (220762_s_at), score: 0.47 GPR144G protein-coupled receptor 144 (216289_at), score: 0.48 GSDMBgasdermin B (215659_at), score: -0.63 HAB1B1 for mucin (215778_x_at), score: 0.66 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.47 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: 0.67 HIST1H2AGhistone cluster 1, H2ag (207156_at), score: -0.6 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: 0.66 HMHB1histocompatibility (minor) HB-1 (208302_at), score: 0.47 HOXA4homeobox A4 (206289_at), score: 0.5 HPNhepsin (204934_s_at), score: 0.61 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: -0.6 HSPA6heat shock 70kDa protein 6 (HSP70B') (117_at), score: 0.64 ICAM2intercellular adhesion molecule 2 (204683_at), score: 0.58 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: 0.55 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.67 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.59 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.49 INHAinhibin, alpha (210141_s_at), score: 0.73 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: -0.49 IRF2interferon regulatory factor 2 (203275_at), score: -0.51 ISOC2isochorismatase domain containing 2 (218893_at), score: 0.57 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.76 JARID1Ajumonji, AT rich interactive domain 1A (215698_at), score: -0.5 KIAA0999KIAA0999 protein (204157_s_at), score: -0.53 KIF16Bkinesin family member 16B (219570_at), score: -0.58 KIF26Bkinesin family member 26B (220002_at), score: 0.52 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: -0.56 KLK13kallikrein-related peptidase 13 (205783_at), score: -0.71 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (205307_s_at), score: -0.77 KRI1KRI1 homolog (S. cerevisiae) (218798_at), score: 0.53 KRT8keratin 8 (209008_x_at), score: 0.58 KRT8P12keratin 8 pseudogene 12 (222060_at), score: 0.79 KSR1kinase suppressor of ras 1 (213769_at), score: 0.59 LATS1LATS, large tumor suppressor, homolog 1 (Drosophila) (219813_at), score: -0.68 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: 0.81 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.63 LILRB4leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4 (210152_at), score: 0.76 LINS1lines homolog 1 (Drosophila) (220121_at), score: -0.5 LOC100128919similar to HSPC157 (219865_at), score: -0.81 LOC100133503similar to LRRC37A3 protein (220219_s_at), score: 0.51 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.57 LOC80054hypothetical LOC80054 (220465_at), score: 0.53 LRBALPS-responsive vesicle trafficking, beach and anchor containing (214109_at), score: -0.75 LTBP4latent transforming growth factor beta binding protein 4 (210628_x_at), score: 0.54 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.72 MAPK8IP2mitogen-activated protein kinase 8 interacting protein 2 (205050_s_at), score: 0.67 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (213178_s_at), score: 0.53 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: -0.75 MED18mediator complex subunit 18 (221650_s_at), score: -0.51 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: 0.51 MGC13053hypothetical MGC13053 (211775_x_at), score: -0.8 MGC5590hypothetical protein MGC5590 (220931_at), score: 0.48 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: -0.55 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.5 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: -0.51 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: 0.52 MUC3Bmucin 3B, cell surface associated (214898_x_at), score: 0.66 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.78 MYBPC1myosin binding protein C, slow type (214087_s_at), score: 0.48 MYL10myosin, light chain 10, regulatory (221659_s_at), score: 0.46 MYO15Amyosin XVA (220288_at), score: 0.63 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.57 NEO1neogenin homolog 1 (chicken) (204321_at), score: -0.52 NEURLneuralized homolog (Drosophila) (204889_s_at), score: 0.51 NINninein (GSK3B interacting protein) (219285_s_at), score: 0.47 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: 0.54 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.48 NPEPL1aminopeptidase-like 1 (89476_r_at), score: 0.55 NUCKS1nuclear casein kinase and cyclin-dependent kinase substrate 1 (217802_s_at), score: 0.53 OPRL1opiate receptor-like 1 (206564_at), score: 0.69 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: 0.47 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: 0.51 PAX4paired box 4 (207867_at), score: -0.62 PCNXL2pecanex-like 2 (Drosophila) (39650_s_at), score: 0.58 PEX13peroxisomal biogenesis factor 13 (205246_at), score: -0.61 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: -0.63 PHF2PHD finger protein 2 (212726_at), score: -0.74 PIAS3protein inhibitor of activated STAT, 3 (203035_s_at), score: -0.56 PIK3R3phosphoinositide-3-kinase, regulatory subunit 3 (gamma) (202743_at), score: -0.55 PIPOXpipecolic acid oxidase (221605_s_at), score: 0.65 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.64 PLLPplasma membrane proteolipid (plasmolipin) (204519_s_at), score: 0.47 PLXNB3plexin B3 (205957_at), score: 0.56 PMS2L2postmeiotic segregation increased 2-like 2 pseudogene (215410_at), score: 0.8 PNMAL1PNMA-like 1 (218824_at), score: -0.57 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.91 PRLHprolactin releasing hormone (221443_x_at), score: 0.52 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: 0.46 PSCAprostate stem cell antigen (205319_at), score: 0.48 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.48 RAB40BRAB40B, member RAS oncogene family (217597_x_at), score: 0.61 RANBP17RAN binding protein 17 (219661_at), score: 0.54 RAP1GAPRAP1 GTPase activating protein (203911_at), score: 0.52 RASGRP2RAS guanyl releasing protein 2 (calcium and DAG-regulated) (214369_s_at), score: 0.59 RBAKRB-associated KRAB zinc finger (219214_s_at), score: 0.49 RBM12BRNA binding motif protein 12B (51228_at), score: -0.82 RBM38RNA binding motif protein 38 (212430_at), score: 0.81 RBM4BRNA binding motif protein 4B (209497_s_at), score: -0.55 RLN1relaxin 1 (211753_s_at), score: 0.67 RNF121ring finger protein 121 (219021_at), score: 0.74 RNF146ring finger protein 146 (221430_s_at), score: 0.61 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: 0.52 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: 0.55 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: 0.5 SARM1sterile alpha and TIR motif containing 1 (213259_s_at), score: 0.59 SCLYselenocysteine lyase (221575_at), score: -0.9 SEPT5septin 5 (209767_s_at), score: 0.75 SHANK2SH3 and multiple ankyrin repeat domains 2 (213307_at), score: -0.78 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.55 SIK2salt-inducible kinase 2 (213221_s_at), score: -0.56 SIX6SIX homeobox 6 (207250_at), score: 0.48 SLC25A21solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 (220474_at), score: -0.56 SLC26A6solute carrier family 26, member 6 (221572_s_at), score: 0.53 SLC27A3solute carrier family 27 (fatty acid transporter), member 3 (222217_s_at), score: -0.52 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: -0.83 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.6 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.75 SNX16sorting nexin 16 (219793_at), score: -0.5 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: 0.53 SPEF1sperm flagellar 1 (216119_s_at), score: 0.61 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.75 STIM1stromal interaction molecule 1 (202764_at), score: -0.52 STXBP2syntaxin binding protein 2 (209367_at), score: 0.66 SUV39H2suppressor of variegation 3-9 homolog 2 (Drosophila) (219262_at), score: -0.72 SYT1synaptotagmin I (203999_at), score: -0.62 SYT5synaptotagmin V (206161_s_at), score: 0.56 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.8 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.52 TAP2transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) (204769_s_at), score: 0.53 TBX10T-box 10 (207689_at), score: 0.57 TCP10t-complex 10 homolog (mouse) (207503_at), score: 0.53 TMEM40transmembrane protein 40 (219503_s_at), score: 0.55 TMPRSS6transmembrane protease, serine 6 (214955_at), score: 0.88 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.55 TNFRSF8tumor necrosis factor receptor superfamily, member 8 (206729_at), score: 0.47 TNIKTRAF2 and NCK interacting kinase (213107_at), score: -0.5 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.61 TP53TG1TP53 target 1 (non-protein coding) (210241_s_at), score: 0.47 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.5 TRAF6TNF receptor-associated factor 6 (205558_at), score: -0.5 TRMT61AtRNA methyltransferase 61 homolog A (S. cerevisiae) (52741_at), score: 0.49 TRPV4transient receptor potential cation channel, subfamily V, member 4 (219516_at), score: 0.57 TSKStestis-specific serine kinase substrate (220545_s_at), score: 0.55 TULP2tubby like protein 2 (206733_at), score: 0.61 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.61 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.47 WFDC8WAP four-disulfide core domain 8 (215276_at), score: 0.54 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.6 ZFP161zinc finger protein 161 homolog (mouse) (208199_s_at), score: 0.55 ZNF132zinc finger protein 132 (207402_at), score: -0.58 ZNF192zinc finger protein 192 (206579_at), score: -0.74 ZNF214zinc finger protein 214 (220497_at), score: -0.7 ZNF33Bzinc finger protein 33B (215022_x_at), score: -0.5 ZNF671zinc finger protein 671 (219849_at), score: 0.48 ZNF767zinc finger family member 767 (219627_at), score: 0.48 ZNF814zinc finger protein 814 (60794_f_at), score: -0.51 ZXDAzinc finger, X-linked, duplicated A (215263_at), score: -0.57
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |