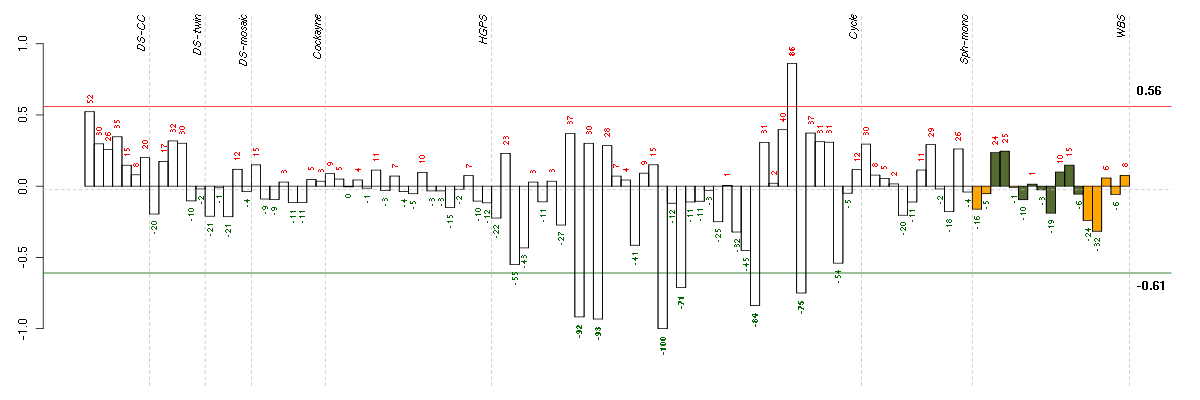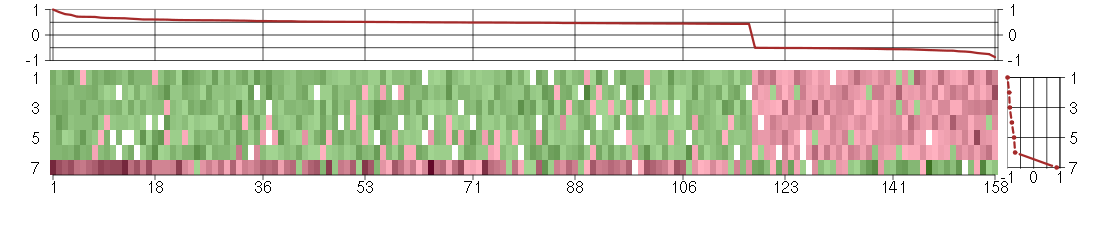

Under-expression is coded with green,
over-expression with red color.

adaptive immune response
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process, and allowing for enhanced response to subsequent exposures to the same antigen (immunological memory).
immune effector process
Any process of the immune system that occurs as part of an immune response.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
leukocyte mediated immunity
Any process involved in the carrying out of an immune response by a leukocyte.
lymphocyte mediated immunity
Any process involved in the carrying out of an immune response by a lymphocyte.
adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process that includes somatic recombination of germline gene segments encoding immunoglobulin superfamily domains, and allowing for enhanced responses upon subsequent exposures to the same antigen (immunological memory). Recombined receptors for antigen encoded by immunoglobulin superfamily domains include T cell receptors and immunoglobulins (antibodies).
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
regulation of immune effector process
Any process that modulates the frequency, rate, or extent of an immune effector process.
positive regulation of immune effector process
Any process that activates or increases the frequency, rate, or extent of an immune effector process.
regulation of leukocyte mediated immunity
Any process that modulates the frequency, rate, or extent of leukocyte mediated immunity.
positive regulation of leukocyte mediated immunity
Any process that activates or increases the frequency, rate, or extent of leukocyte mediated immunity.
regulation of lymphocyte mediated immunity
Any process that modulates the frequency, rate, or extent of lymphocyte mediated immunity.
positive regulation of lymphocyte mediated immunity
Any process that activates or increases the frequency, rate, or extent of lymphocyte mediated immunity.
regulation of B cell mediated immunity
Any process that modulates the frequency, rate, or extent of B cell mediated immunity.
positive regulation of B cell mediated immunity
Any process that activates or increases the frequency, rate, or extent of B cell mediated immunity.
regulation of adaptive immune response
Any process that modulates the frequency, rate, or extent of an adaptive immune response.
positive regulation of adaptive immune response
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response.
regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
Any process that modulates the frequency, rate, or extent of an adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains.
positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains.
regulation of immunoglobulin mediated immune response
Any process that modulates the frequency, rate, or extent of an immunoglobulin mediated immune response.
positive regulation of immunoglobulin mediated immune response
Any process that activates or increases the frequency, rate, or extent of an immunoglobulin mediated immune response.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
immunoglobulin mediated immune response
An immune response mediated by immunoglobulins, whether cell-bound or in solution.
B cell mediated immunity
Any process involved with the carrying out of an immune response by a B cell, through, for instance, the production of antibodies or cytokines, or antigen presentation to T cells.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
positive regulation of immune effector process
Any process that activates or increases the frequency, rate, or extent of an immune effector process.
immune effector process
Any process of the immune system that occurs as part of an immune response.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune effector process
Any process that activates or increases the frequency, rate, or extent of an immune effector process.
regulation of leukocyte mediated immunity
Any process that modulates the frequency, rate, or extent of leukocyte mediated immunity.
positive regulation of leukocyte mediated immunity
Any process that activates or increases the frequency, rate, or extent of leukocyte mediated immunity.
regulation of immune effector process
Any process that modulates the frequency, rate, or extent of an immune effector process.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
regulation of adaptive immune response
Any process that modulates the frequency, rate, or extent of an adaptive immune response.
positive regulation of adaptive immune response
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response.
positive regulation of leukocyte mediated immunity
Any process that activates or increases the frequency, rate, or extent of leukocyte mediated immunity.
regulation of lymphocyte mediated immunity
Any process that modulates the frequency, rate, or extent of lymphocyte mediated immunity.
positive regulation of lymphocyte mediated immunity
Any process that activates or increases the frequency, rate, or extent of lymphocyte mediated immunity.
positive regulation of adaptive immune response
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response.
regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
Any process that modulates the frequency, rate, or extent of an adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains.
positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains.
B cell mediated immunity
Any process involved with the carrying out of an immune response by a B cell, through, for instance, the production of antibodies or cytokines, or antigen presentation to T cells.
positive regulation of lymphocyte mediated immunity
Any process that activates or increases the frequency, rate, or extent of lymphocyte mediated immunity.
regulation of B cell mediated immunity
Any process that modulates the frequency, rate, or extent of B cell mediated immunity.
positive regulation of B cell mediated immunity
Any process that activates or increases the frequency, rate, or extent of B cell mediated immunity.
regulation of B cell mediated immunity
Any process that modulates the frequency, rate, or extent of B cell mediated immunity.
positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains.
positive regulation of B cell mediated immunity
Any process that activates or increases the frequency, rate, or extent of B cell mediated immunity.
positive regulation of B cell mediated immunity
Any process that activates or increases the frequency, rate, or extent of B cell mediated immunity.
regulation of immunoglobulin mediated immune response
Any process that modulates the frequency, rate, or extent of an immunoglobulin mediated immune response.
positive regulation of immunoglobulin mediated immune response
Any process that activates or increases the frequency, rate, or extent of an immunoglobulin mediated immune response.
positive regulation of immunoglobulin mediated immune response
Any process that activates or increases the frequency, rate, or extent of an immunoglobulin mediated immune response.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

ABCG2ATP-binding cassette, sub-family G (WHITE), member 2 (209735_at), score: -0.64 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.5 AFF2AF4/FMR2 family, member 2 (206105_at), score: 0.48 AGAP2ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 (215080_s_at), score: 0.46 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.48 APBA2amyloid beta (A4) precursor protein-binding, family A, member 2 (209871_s_at), score: 0.46 ARHGAP8Rho GTPase activating protein 8 (37117_at), score: 0.53 ARHGDIBRho GDP dissociation inhibitor (GDI) beta (201288_at), score: -0.5 ASPNasporin (219087_at), score: 0.59 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: -0.57 B9D1B9 protein domain 1 (210534_s_at), score: -0.51 BAHD1bromo adjacent homology domain containing 1 (203051_at), score: 0.49 BAI2brain-specific angiogenesis inhibitor 2 (204966_at), score: 0.61 BEX1brain expressed, X-linked 1 (218332_at), score: -0.73 C10orf59chromosome 10 open reading frame 59 (220564_at), score: 0.52 C10orf72chromosome 10 open reading frame 72 (213381_at), score: 0.49 C11orf67chromosome 11 open reading frame 67 (221600_s_at), score: -0.58 C3complement component 3 (217767_at), score: 0.52 CACNA1Gcalcium channel, voltage-dependent, T type, alpha 1G subunit (211315_s_at), score: 0.71 CAMK2Bcalcium/calmodulin-dependent protein kinase II beta (34846_at), score: 0.47 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: 1 CD226CD226 molecule (207315_at), score: 0.5 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213348_at), score: 0.49 CEACAM1carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) (211883_x_at), score: 0.57 CEP70centrosomal protein 70kDa (219036_at), score: -0.51 CFBcomplement factor B (202357_s_at), score: 0.54 CHRDL1chordin-like 1 (209763_at), score: 0.55 CHRNA6cholinergic receptor, nicotinic, alpha 6 (207568_at), score: 0.48 CHST1carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 (205567_at), score: 0.46 CLEC3BC-type lectin domain family 3, member B (205200_at), score: 0.64 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: 0.61 COL21A1collagen, type XXI, alpha 1 (208096_s_at), score: 0.66 COL5A3collagen, type V, alpha 3 (52255_s_at), score: 0.58 COQ2coenzyme Q2 homolog, prenyltransferase (yeast) (213379_at), score: -0.51 COX15COX15 homolog, cytochrome c oxidase assembly protein (yeast) (221550_at), score: -0.53 CPA3carboxypeptidase A3 (mast cell) (205624_at), score: 0.45 CPEB3cytoplasmic polyadenylation element binding protein 3 (205773_at), score: 0.61 CPZcarboxypeptidase Z (211062_s_at), score: 0.51 CSF2RBcolony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) (205159_at), score: 0.46 CSTF2cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa (204459_at), score: -0.59 CTSDcathepsin D (200766_at), score: 0.46 CTSFcathepsin F (203657_s_at), score: 0.5 CTSOcathepsin O (203758_at), score: 0.48 CXorf15chromosome X open reading frame 15 (219969_at), score: 0.49 CYorf14chromosome Y open reading frame 14 (207063_at), score: 0.56 CYP3A43cytochrome P450, family 3, subfamily A, polypeptide 43 (211440_x_at), score: 0.82 DHCR77-dehydrocholesterol reductase (201790_s_at), score: 0.5 DHRS2dehydrogenase/reductase (SDR family) member 2 (214079_at), score: 0.52 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.49 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.52 DNASE1L3deoxyribonuclease I-like 3 (205554_s_at), score: 0.71 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: 0.47 ECM2extracellular matrix protein 2, female organ and adipocyte specific (206101_at), score: 0.57 EGFL6EGF-like-domain, multiple 6 (219454_at), score: 0.51 EMCNendomucin (219436_s_at), score: -0.87 EPHB6EPH receptor B6 (204718_at), score: 0.51 EVI2Becotropic viral integration site 2B (211742_s_at), score: -0.53 F2RL1coagulation factor II (thrombin) receptor-like 1 (213506_at), score: -0.51 FA2Hfatty acid 2-hydroxylase (219429_at), score: 0.72 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: 0.71 FADS2fatty acid desaturase 2 (202218_s_at), score: 0.5 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: 0.45 FCGRTFc fragment of IgG, receptor, transporter, alpha (218831_s_at), score: 0.45 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: 0.46 G6PC2glucose-6-phosphatase, catalytic, 2 (221453_at), score: 0.52 GAAglucosidase, alpha; acid (202812_at), score: 0.67 GABRG3gamma-aminobutyric acid (GABA) A receptor, gamma 3 (216895_at), score: 0.47 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.48 GK2glycerol kinase 2 (215430_at), score: 0.51 GPR4G protein-coupled receptor 4 (206236_at), score: 0.45 GPR56G protein-coupled receptor 56 (212070_at), score: -0.56 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: -0.51 GRAMD1CGRAM domain containing 1C (219313_at), score: -0.54 GRPgastrin-releasing peptide (206326_at), score: -0.52 H6PDhexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (221892_at), score: 0.55 HIST1H1Chistone cluster 1, H1c (209398_at), score: -0.52 HIST1H4Ghistone cluster 1, H4g (208551_at), score: 0.51 HLA-DPA1major histocompatibility complex, class II, DP alpha 1 (211990_at), score: 0.63 HOXD1homeobox D1 (205975_s_at), score: 0.44 HSD17B2hydroxysteroid (17-beta) dehydrogenase 2 (204818_at), score: 0.45 IFI30interferon, gamma-inducible protein 30 (201422_at), score: -0.52 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: 0.66 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.58 KCND3potassium voltage-gated channel, Shal-related subfamily, member 3 (213832_at), score: 0.47 KIAA0644KIAA0644 gene product (205150_s_at), score: 0.57 L3MBTLl(3)mbt-like (Drosophila) (213837_at), score: 0.48 LOC729806similar to hCG1725380 (217544_at), score: 0.44 LPCAT3lysophosphatidylcholine acyltransferase 3 (202793_at), score: 0.49 LY6Hlymphocyte antigen 6 complex, locus H (206773_at), score: 0.44 MAGEB2melanoma antigen family B, 2 (206218_at), score: 0.9 MAN2A2mannosidase, alpha, class 2A, member 2 (219999_at), score: 0.44 MAPK10mitogen-activated protein kinase 10 (204813_at), score: 0.44 MCF2L2MCF.2 cell line derived transforming sequence-like 2 (215112_x_at), score: 0.44 MEGF6multiple EGF-like-domains 6 (213942_at), score: 0.52 MEP1Bmeprin A, beta (207251_at), score: 0.49 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: 0.47 MLYCDmalonyl-CoA decarboxylase (218869_at), score: -0.61 MMEmembrane metallo-endopeptidase (203434_s_at), score: 0.44 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.49 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: -0.56 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: 0.47 MUC13mucin 13, cell surface associated (218687_s_at), score: 0.52 MYL5myosin, light chain 5, regulatory (205145_s_at), score: 0.45 MYO5Cmyosin VC (218966_at), score: 0.44 NAIPNLR family, apoptosis inhibitory protein (204860_s_at), score: 0.59 NDRG4NDRG family member 4 (209159_s_at), score: -0.52 NEFLneurofilament, light polypeptide (221805_at), score: 0.58 NEK11NIMA (never in mitosis gene a)- related kinase 11 (219542_at), score: 0.48 NOS1nitric oxide synthase 1 (neuronal) (207309_at), score: 0.47 NOVA1neuro-oncological ventral antigen 1 (205794_s_at), score: 0.6 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: -0.54 OCLMoculomedin (208274_at), score: 0.5 OLFML2Aolfactomedin-like 2A (213075_at), score: 0.79 OLFML2Bolfactomedin-like 2B (213125_at), score: -0.61 OMDosteomodulin (205907_s_at), score: 0.55 P2RY4pyrimidinergic receptor P2Y, G-protein coupled, 4 (221466_at), score: 0.5 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.6 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.55 PDGFDplatelet derived growth factor D (219304_s_at), score: 0.54 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: -0.51 PLXDC1plexin domain containing 1 (219700_at), score: 0.58 PPLperiplakin (203407_at), score: 0.49 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: 0.68 PTPN20Bprotein tyrosine phosphatase, non-receptor type 20B (215172_at), score: -0.65 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: 0.54 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -0.56 RETret proto-oncogene (205879_x_at), score: 0.45 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: 0.52 S100A3S100 calcium binding protein A3 (206027_at), score: -0.51 SAA4serum amyloid A4, constitutive (207096_at), score: 0.48 SEMA3Csema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C (203788_s_at), score: 0.58 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: 0.52 SLAMF1signaling lymphocytic activation molecule family member 1 (206181_at), score: 0.6 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: 0.45 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: 0.55 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.5 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: -0.75 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (212947_at), score: 0.45 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.71 SPA17sperm autoantigenic protein 17 (205406_s_at), score: -0.55 SPAG6sperm associated antigen 6 (210033_s_at), score: 0.46 SYCP2synaptonemal complex protein 2 (206546_at), score: -0.5 TAF4BTAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa (216226_at), score: 0.49 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: -0.52 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.53 TM7SF2transmembrane 7 superfamily member 2 (210130_s_at), score: 0.66 TMSB15Athymosin beta 15a (205347_s_at), score: -0.61 TNFSF10tumor necrosis factor (ligand) superfamily, member 10 (214329_x_at), score: 0.56 TNFSF9tumor necrosis factor (ligand) superfamily, member 9 (206907_at), score: 0.52 TNIKTRAF2 and NCK interacting kinase (213107_at), score: -0.56 TOP3Btopoisomerase (DNA) III beta (215781_s_at), score: -0.51 TP63tumor protein p63 (209863_s_at), score: 0.51 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: 0.44 TSKStestis-specific serine kinase substrate (220545_s_at), score: 0.49 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: -0.67 ZNF117zinc finger protein 117 (207605_x_at), score: 0.54 ZNF219zinc finger protein 219 (219314_s_at), score: 0.58 ZNF3zinc finger protein 3 (212684_at), score: -0.54
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |