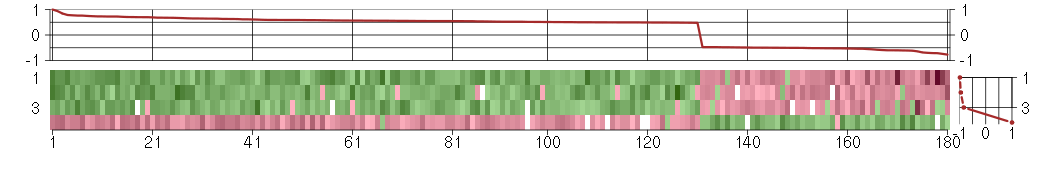

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
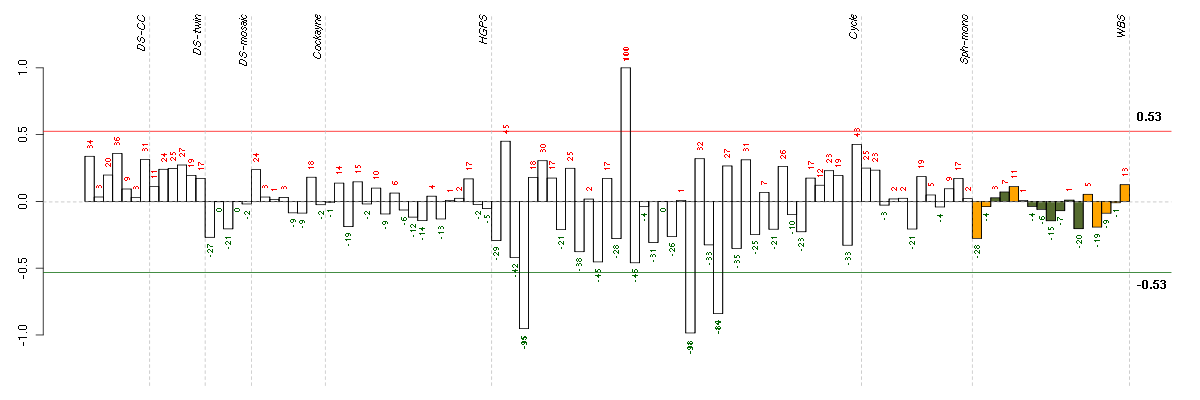
ABCC9ATP-binding cassette, sub-family C (CFTR/MRP), member 9 (208561_at), score: -0.52 ACADSacyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain (202366_at), score: 0.57 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.51 ADORA1adenosine A1 receptor (216220_s_at), score: 0.56 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.51 APPamyloid beta (A4) precursor protein (214953_s_at), score: 0.68 ARHGEF10LRho guanine nucleotide exchange factor (GEF) 10-like (221656_s_at), score: 0.55 ASB13ankyrin repeat and SOCS box-containing 13 (218862_at), score: 0.5 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214244_s_at), score: 0.65 B3GALT4UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (210205_at), score: 0.6 BAGEB melanoma antigen (207712_at), score: -0.61 BAIAP3BAI1-associated protein 3 (216356_x_at), score: 0.55 BCANbrevican (91920_at), score: 0.55 BEST1bestrophin 1 (207671_s_at), score: 0.49 C10orf110chromosome 10 open reading frame 110 (220703_at), score: 0.5 C10orf81chromosome 10 open reading frame 81 (219857_at), score: -0.71 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: -0.52 C3orf51chromosome 3 open reading frame 51 (208247_at), score: 0.53 C4Acomplement component 4A (Rodgers blood group) (214428_x_at), score: 0.65 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.51 C8orf60chromosome 8 open reading frame 60 (220712_at), score: 0.59 CALB1calbindin 1, 28kDa (205626_s_at), score: 0.57 CCBP2chemokine binding protein 2 (206887_at), score: 0.49 CCR1chemokine (C-C motif) receptor 1 (205099_s_at), score: 0.52 CD53CD53 molecule (203416_at), score: 0.65 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.7 CHPcalcium binding protein P22 (214665_s_at), score: 0.51 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: 0.61 CNNM3cyclin M3 (220739_s_at), score: -0.48 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.7 COL9A2collagen, type IX, alpha 2 (213622_at), score: 0.75 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.5 COX16COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) (217645_at), score: 0.52 CPA2carboxypeptidase A2 (pancreatic) (206212_at), score: 0.52 CRKRSCdc2-related kinase, arginine/serine-rich (219226_at), score: -0.53 CSF2RBcolony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) (205159_at), score: -0.49 CTTNcortactin (201059_at), score: 0.54 CYTH4cytohesin 4 (219183_s_at), score: 0.52 DOK4docking protein 4 (209691_s_at), score: -0.56 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.84 EDAectodysplasin A (211130_x_at), score: 0.71 EGLN3egl nine homolog 3 (C. elegans) (219232_s_at), score: -0.71 F7coagulation factor VII (serum prothrombin conversion accelerator) (207300_s_at), score: 0.53 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: 0.67 FBRSfibrosin (218255_s_at), score: 0.78 FBXO2F-box protein 2 (219305_x_at), score: 0.64 FETUBfetuin B (214417_s_at), score: 0.54 FGL1fibrinogen-like 1 (205305_at), score: 0.73 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.71 FLJ21075hypothetical protein FLJ21075 (221172_at), score: 0.49 FNTBfarnesyltransferase, CAAX box, beta (204764_at), score: -0.47 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: 0.57 GANgigaxonin (220124_at), score: -0.5 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: 0.68 GGTLC1gamma-glutamyltransferase light chain 1 (211416_x_at), score: 0.5 GPATCH4G patch domain containing 4 (220596_at), score: 0.5 GPR144G protein-coupled receptor 144 (216289_at), score: 0.72 GPR32G protein-coupled receptor 32 (221469_at), score: 0.49 GRM1glutamate receptor, metabotropic 1 (210939_s_at), score: 0.58 HAB1B1 for mucin (215778_x_at), score: 0.77 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.7 HPNhepsin (204934_s_at), score: 0.55 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.48 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (217039_x_at), score: 0.61 IL18R1interleukin 18 receptor 1 (206618_at), score: -0.5 IL22RA1interleukin 22 receptor, alpha 1 (220056_at), score: 0.51 ISOC2isochorismatase domain containing 2 (218893_at), score: 0.59 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.94 ITGB3integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) (204628_s_at), score: 0.49 JAG2jagged 2 (32137_at), score: 0.55 KCTD7potassium channel tetramerisation domain containing 7 (213474_at), score: 0.5 KIAA0355KIAA0355 (203288_at), score: -0.52 KIAA0652KIAA0652 (203363_s_at), score: -0.5 KLHL4kelch-like 4 (Drosophila) (214591_at), score: -0.7 KRT24keratin 24 (220267_at), score: 0.57 KRT86keratin 86 (215189_at), score: 0.53 KRT9keratin 9 (208188_at), score: 0.49 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.63 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.55 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.74 LOC391132similar to hCG2041276 (216177_at), score: 0.59 LOC80054hypothetical LOC80054 (220465_at), score: 0.59 LRRC37A2leucine rich repeat containing 37, member A2 (221740_x_at), score: 0.57 MAGOH2mago-nashi homolog 2, proliferation-associated (Drosophila) (217693_x_at), score: 0.52 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.52 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.51 MATN3matrilin 3 (206091_at), score: -0.74 MEOX1mesenchyme homeobox 1 (205619_s_at), score: 0.69 MGAT4Cmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) (207447_s_at), score: -0.64 MUC3Bmucin 3B, cell surface associated (214898_x_at), score: 0.55 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.55 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: -0.48 N4BP3Nedd4 binding protein 3 (214775_at), score: 0.51 NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: 0.76 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.55 NDST1N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 (202608_s_at), score: -0.51 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: 0.48 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.48 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: -0.48 OPRL1opiate receptor-like 1 (206564_at), score: 0.55 PCDH24protocadherin 24 (220186_s_at), score: 0.5 PCSK5proprotein convertase subtilisin/kexin type 5 (213652_at), score: -0.48 PDE8Bphosphodiesterase 8B (213228_at), score: 0.49 PDK3pyruvate dehydrogenase kinase, isozyme 3 (221957_at), score: -0.68 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.73 PHC3polyhomeotic homolog 3 (Drosophila) (215521_at), score: 0.63 PIAS4protein inhibitor of activated STAT, 4 (212881_at), score: 0.55 PIN1Lpeptidylprolyl cis/trans isomerase, NIMA-interacting 1-like (pseudogene) (207582_at), score: 0.5 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.63 PKN1protein kinase N1 (202161_at), score: -0.48 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.56 PPP1R10protein phosphatase 1, regulatory (inhibitor) subunit 10 (201702_s_at), score: -0.5 PPP1R9Aprotein phosphatase 1, regulatory (inhibitor) subunit 9A (221088_s_at), score: -0.77 PRAMEF11PRAME family member 11 (217365_at), score: 0.51 PRLHprolactin releasing hormone (221443_x_at), score: 0.55 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 1 PXNpaxillin (211823_s_at), score: -0.6 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.72 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -0.53 RFX3regulatory factor X, 3 (influences HLA class II expression) (207234_at), score: -0.6 RHEBRas homolog enriched in brain (213409_s_at), score: 0.51 RICH2Rho-type GTPase-activating protein RICH2 (215232_at), score: 0.59 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: 0.56 RLN1relaxin 1 (211753_s_at), score: 0.58 RNF126P1ring finger protein 126 pseudogene 1 (222349_x_at), score: 0.5 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: 0.49 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.56 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: 0.49 RPL27Aribosomal protein L27a (212044_s_at), score: 0.58 RPL38ribosomal protein L38 (202028_s_at), score: 0.6 S100A14S100 calcium binding protein A14 (218677_at), score: 0.63 SAA4serum amyloid A4, constitutive (207096_at), score: 0.64 SARM1sterile alpha and TIR motif containing 1 (213259_s_at), score: 0.5 SEC14L5SEC14-like 5 (S. cerevisiae) (210169_at), score: 0.56 SEPT5septin 5 (209767_s_at), score: 0.56 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.59 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.61 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: -0.49 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (210686_x_at), score: 0.48 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.6 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: -0.5 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: 0.65 SLC9A7solute carrier family 9 (sodium/hydrogen exchanger), member 7 (214860_at), score: -0.5 SNHG3small nucleolar RNA host gene 3 (non-protein coding) (215011_at), score: 0.54 SPATA1spermatogenesis associated 1 (221057_at), score: 0.66 SPATA6spermatogenesis associated 6 (220298_s_at), score: 0.58 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: -0.52 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.56 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.59 STIM1stromal interaction molecule 1 (202764_at), score: -0.62 SULT1A2sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (207122_x_at), score: 0.51 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.68 TAOK2TAO kinase 2 (204986_s_at), score: -0.55 TBX21T-box 21 (220684_at), score: 0.49 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: 0.56 TMC5transmembrane channel-like 5 (219580_s_at), score: 0.53 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: 0.68 TMEM62transmembrane protein 62 (218776_s_at), score: -0.54 TMEM92transmembrane protein 92 (216791_at), score: 0.48 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.72 TNFRSF14tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) (209354_at), score: 0.58 TNFRSF8tumor necrosis factor receptor superfamily, member 8 (206729_at), score: 0.76 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.64 TNK2tyrosine kinase, non-receptor, 2 (203839_s_at), score: -0.54 TSPAN12tetraspanin 12 (219274_at), score: -0.49 TULP2tubby like protein 2 (206733_at), score: 0.7 TXNthioredoxin (216609_at), score: 0.52 TYMPthymidine phosphorylase (217497_at), score: 0.49 TYRO3TYRO3 protein tyrosine kinase (211432_s_at), score: -0.5 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.51 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.56 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.5 WDR52WD repeat domain 52 (221103_s_at), score: -0.61 WDTC1WD and tetratricopeptide repeats 1 (215497_s_at), score: 0.59 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: 0.49 ZNF224zinc finger protein 224 (216983_s_at), score: 0.49 ZNF318zinc finger protein 318 (203521_s_at), score: -0.52 ZNF334zinc finger protein 334 (220022_at), score: -0.58 ZNF516zinc finger protein 516 (203604_at), score: 0.5 ZNF814zinc finger protein 814 (60794_f_at), score: -0.59
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |