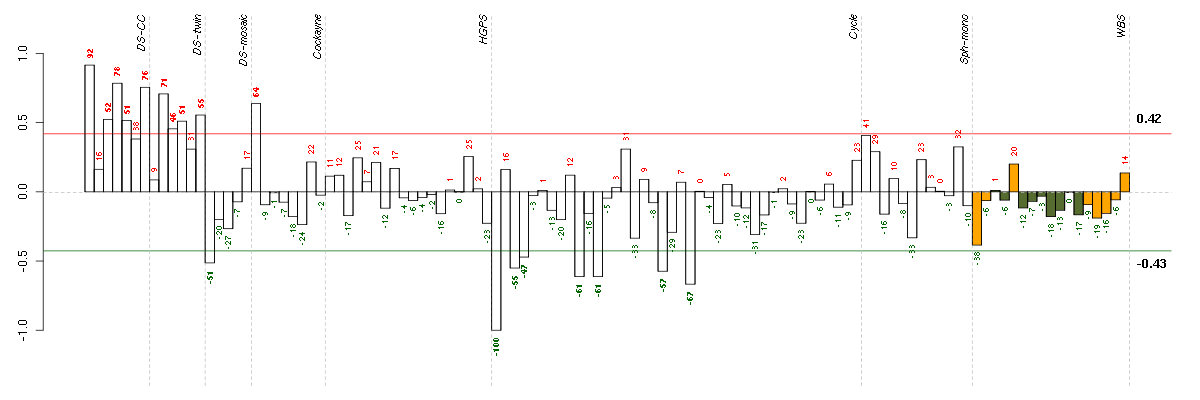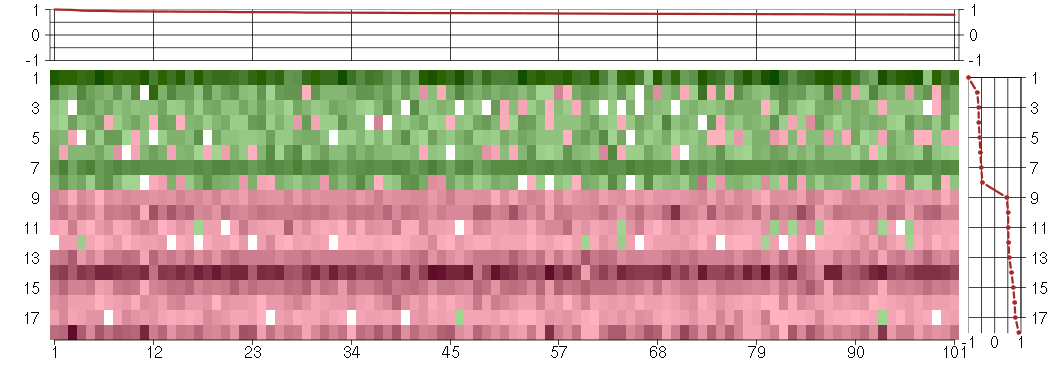

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
all
This term is the most general term possible

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

A1CFAPOBEC1 complementation factor (220951_s_at), score: 0.96 ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: 0.82 AFF2AF4/FMR2 family, member 2 (206105_at), score: 0.83 AGMATagmatine ureohydrolase (agmatinase) (219792_at), score: 0.88 AIPL1aryl hydrocarbon receptor interacting protein-like 1 (219977_at), score: 0.86 APOMapolipoprotein M (205682_x_at), score: 0.85 BMP10bone morphogenetic protein 10 (208292_at), score: 0.83 BTNL3butyrophilin-like 3 (217207_s_at), score: 1 C1orf61chromosome 1 open reading frame 61 (205103_at), score: 0.92 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.83 C8orf17chromosome 8 open reading frame 17 (208266_at), score: 0.86 C8orf60chromosome 8 open reading frame 60 (220712_at), score: 0.89 CA4carbonic anhydrase IV (206209_s_at), score: 0.81 CASZ1castor zinc finger 1 (220015_at), score: 0.86 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.82 CD53CD53 molecule (203416_at), score: 0.88 CEACAM5carcinoembryonic antigen-related cell adhesion molecule 5 (201884_at), score: 0.84 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: 0.82 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (220446_s_at), score: 0.9 CLDN10claudin 10 (205328_at), score: 0.9 CLEC4MC-type lectin domain family 4, member M (207995_s_at), score: 0.84 CLEC7AC-type lectin domain family 7, member A (221698_s_at), score: 0.84 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.91 CTSEcathepsin E (205927_s_at), score: 0.9 CYBBcytochrome b-245, beta polypeptide (203923_s_at), score: 0.86 CYP2A6cytochrome P450, family 2, subfamily A, polypeptide 6 (211295_x_at), score: 0.91 CYP2C9cytochrome P450, family 2, subfamily C, polypeptide 9 (214421_x_at), score: 0.8 DHRS9dehydrogenase/reductase (SDR family) member 9 (219799_s_at), score: 0.86 DNASE1L3deoxyribonuclease I-like 3 (205554_s_at), score: 0.91 DRD5dopamine receptor D5 (208486_at), score: 0.9 EDARectodysplasin A receptor (220048_at), score: 0.89 FAM38Bfamily with sequence similarity 38, member B (219602_s_at), score: 0.87 FGL1fibrinogen-like 1 (205305_at), score: 0.87 FOLH1folate hydrolase (prostate-specific membrane antigen) 1 (217487_x_at), score: 0.8 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.83 GNRHRgonadotropin-releasing hormone receptor (216341_s_at), score: 0.81 GPATCH4G patch domain containing 4 (220596_at), score: 0.79 GPR12G protein-coupled receptor 12 (214558_at), score: 0.83 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: 0.85 HAB1B1 for mucin (215778_x_at), score: 0.83 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: 0.82 HIST1H4Dhistone cluster 1, H4d (208076_at), score: 0.89 HOXD4homeobox D4 (205522_at), score: 0.8 HS3ST2heparan sulfate (glucosamine) 3-O-sulfotransferase 2 (219697_at), score: 0.82 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.83 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.84 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (219287_at), score: 0.81 KCNQ3potassium voltage-gated channel, KQT-like subfamily, member 3 (206573_at), score: 0.81 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: 0.87 KLF1Kruppel-like factor 1 (erythroid) (210504_at), score: 0.91 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (205307_s_at), score: 0.86 KRT2keratin 2 (207908_at), score: 0.9 LHX3LIM homeobox 3 (221670_s_at), score: 0.91 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: 0.92 LONRF3LON peptidase N-terminal domain and ring finger 3 (220009_at), score: 0.81 MALmal, T-cell differentiation protein (204777_s_at), score: 0.87 MBmyoglobin (204179_at), score: 0.84 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: 0.8 MMP9matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (203936_s_at), score: 0.83 MMRN2multimerin 2 (219091_s_at), score: 0.85 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.95 MPP3membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) (206186_at), score: 0.81 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: 0.83 MUC3Amucin 3A, cell surface associated (217117_x_at), score: 0.86 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.87 MYO1Amyosin IA (211916_s_at), score: 0.8 MYO5Cmyosin VC (218966_at), score: 0.84 N4BP3Nedd4 binding protein 3 (214775_at), score: 0.94 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: 0.82 NEUROD2neurogenic differentiation 2 (210271_at), score: 0.81 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: 0.8 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.86 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: 0.93 OVOL2ovo-like 2 (Drosophila) (211778_s_at), score: 0.82 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.99 PEG3paternally expressed 3 (209242_at), score: 0.85 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.87 PPP4R4protein phosphatase 4, regulatory subunit 4 (220672_at), score: 0.82 PRB3proline-rich protein BstNI subfamily 3 (206998_x_at), score: 0.87 PRKCBprotein kinase C, beta (209685_s_at), score: 0.82 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: 0.93 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.93 RETret proto-oncogene (205879_x_at), score: 0.95 RICH2Rho-type GTPase-activating protein RICH2 (215232_at), score: 0.8 RP11-35N6.1plasticity related gene 3 (219732_at), score: 0.86 RP3-377H14.5hypothetical LOC285830 (222279_at), score: 0.82 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 0.93 RPH3Arabphilin 3A homolog (mouse) (205230_at), score: 0.8 S100A14S100 calcium binding protein A14 (218677_at), score: 0.91 SCN10Asodium channel, voltage-gated, type X, alpha subunit (208578_at), score: 0.86 SERPINA1serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (211429_s_at), score: 0.92 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: 0.88 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: 0.83 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: 0.86 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.88 SPARCL1SPARC-like 1 (hevin) (200795_at), score: 0.8 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: 0.86 TBX21T-box 21 (220684_at), score: 0.83 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.81 TP63tumor protein p63 (209863_s_at), score: 0.85 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.98
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |