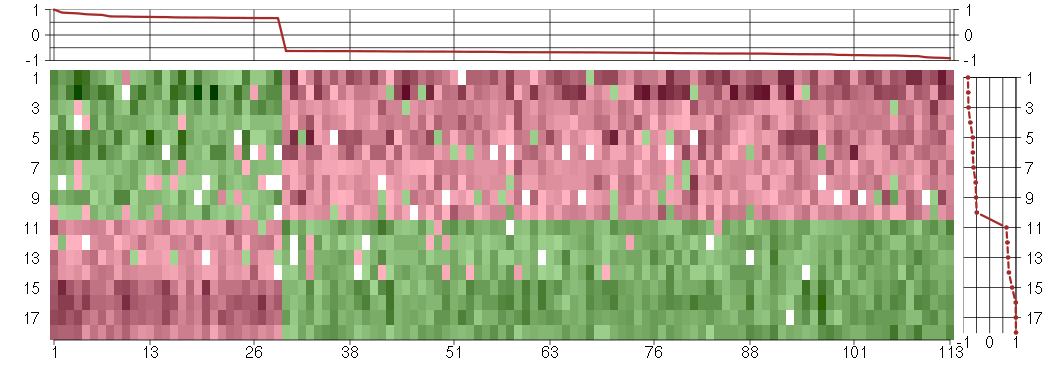

Under-expression is coded with green,
over-expression with red color.

cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
homophilic cell adhesion
The attachment of an adhesion molecule in one cell to an identical molecule in an adjacent cell.
calcium-dependent cell-cell adhesion
The attachment of one cell to another cell via adhesion molecules that require the presence of calcium for the interaction.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell-cell adhesion
The attachment of one cell to another cell via adhesion molecules.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
all
This term is the most general term possible
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.


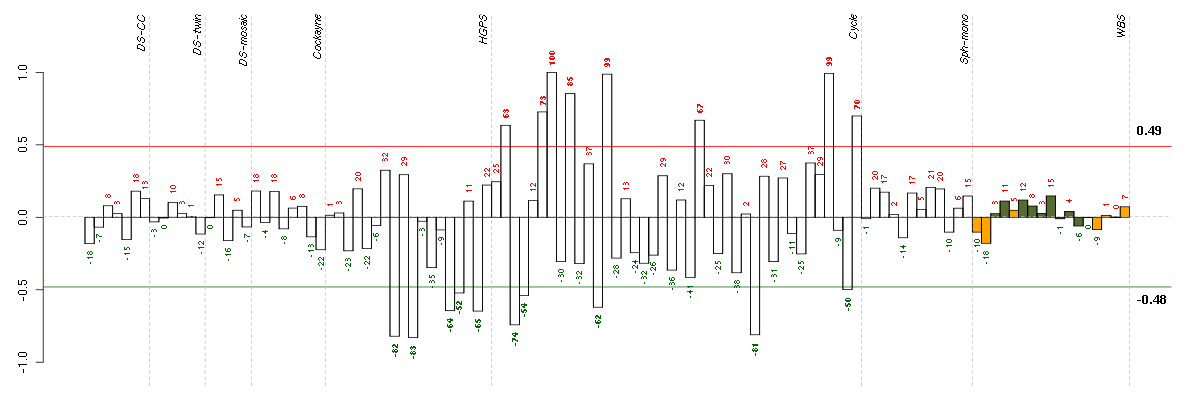
ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: -0.66 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: -0.66 ATN1atrophin 1 (40489_at), score: -0.69 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.69 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: -0.73 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: -0.74 BAIAP2BAI1-associated protein 2 (209502_s_at), score: -0.72 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: -0.65 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.69 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.73 C20orf117chromosome 20 open reading frame 117 (207711_at), score: -0.64 C20orf20chromosome 20 open reading frame 20 (218586_at), score: -0.63 C4orf43chromosome 4 open reading frame 43 (218513_at), score: 0.66 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: -0.65 CASC3cancer susceptibility candidate 3 (207842_s_at), score: -0.63 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: -0.68 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.79 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: 0.79 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: 0.7 CDKN2Dcyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) (210240_s_at), score: -0.63 CICcapicua homolog (Drosophila) (212784_at), score: -0.76 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.81 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: -0.8 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: -0.79 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.8 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.76 CScitrate synthase (208660_at), score: -0.67 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.7 DNM1dynamin 1 (215116_s_at), score: -0.72 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: 0.67 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.83 FOSL1FOS-like antigen 1 (204420_at), score: -0.68 FOXK2forkhead box K2 (203064_s_at), score: -0.74 FOXM1forkhead box M1 (202580_x_at), score: -0.64 FREQfrequenin homolog (Drosophila) (218266_s_at), score: -0.63 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: 0.66 H1FXH1 histone family, member X (204805_s_at), score: -0.75 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: -0.68 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.75 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.68 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: -0.78 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.67 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.8 ITGA1integrin, alpha 1 (214660_at), score: 0.67 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: 0.71 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.88 LIG1ligase I, DNA, ATP-dependent (202726_at), score: -0.69 LMNB2lamin B2 (216952_s_at), score: -0.66 LOC100132540similar to LOC339047 protein (214870_x_at), score: -0.67 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.71 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.65 LRFN4leucine rich repeat and fibronectin type III domain containing 4 (219491_at), score: -0.65 LRRC15leucine rich repeat containing 15 (213909_at), score: -0.63 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: 0.85 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.7 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.68 MAP7D1MAP7 domain containing 1 (217943_s_at), score: -0.68 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: -0.63 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: -0.7 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.86 MYO1Bmyosin IB (212364_at), score: 0.68 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.9 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.72 NEFMneurofilament, medium polypeptide (205113_at), score: 1 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.66 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.73 NRXN3neurexin 3 (205795_at), score: 0.81 OLFML2Bolfactomedin-like 2B (213125_at), score: -0.89 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.67 PARVBparvin, beta (204629_at), score: -0.73 PCDH9protocadherin 9 (219737_s_at), score: 0.72 PCDHG@protocadherin gamma cluster (215836_s_at), score: -0.65 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.73 PCDHGC3protocadherin gamma subfamily C, 3 (211066_x_at), score: -0.63 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.73 PKN1protein kinase N1 (202161_at), score: -0.63 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: -0.65 POLSpolymerase (DNA directed) sigma (202466_at), score: -0.71 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.78 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: -0.64 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: -0.68 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.71 RAB1ARAB1A, member RAS oncogene family (213440_at), score: 0.67 RNF11ring finger protein 11 (208924_at), score: 0.69 RNF220ring finger protein 220 (219988_s_at), score: -0.8 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.8 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.73 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: -0.63 SAP30LSAP30-like (219129_s_at), score: -0.64 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.91 SCARA3scavenger receptor class A, member 3 (219416_at), score: -0.72 SDC1syndecan 1 (201287_s_at), score: -0.76 SF1splicing factor 1 (208313_s_at), score: -0.87 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.68 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: -0.66 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.83 STIM1stromal interaction molecule 1 (202764_at), score: -0.69 STRN4striatin, calmodulin binding protein 4 (217903_at), score: -0.65 SUSD5sushi domain containing 5 (214954_at), score: 0.69 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: 0.72 TBCDtubulin folding cofactor D (211052_s_at), score: -0.68 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.67 TMSB15Bthymosin beta 15B (214051_at), score: -0.69 TXLNAtaxilin alpha (212300_at), score: -0.68 UGDHUDP-glucose dehydrogenase (203343_at), score: 0.66 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.65 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.7 WDR6WD repeat domain 6 (217734_s_at), score: -0.68 WDR62WD repeat domain 62 (215218_s_at), score: -0.73 WHSC1Wolf-Hirschhorn syndrome candidate 1 (209053_s_at), score: -0.63 YTHDC2YTH domain containing 2 (213077_at), score: 0.71 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: -0.68 ZNF536zinc finger protein 536 (206403_at), score: -0.75
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |