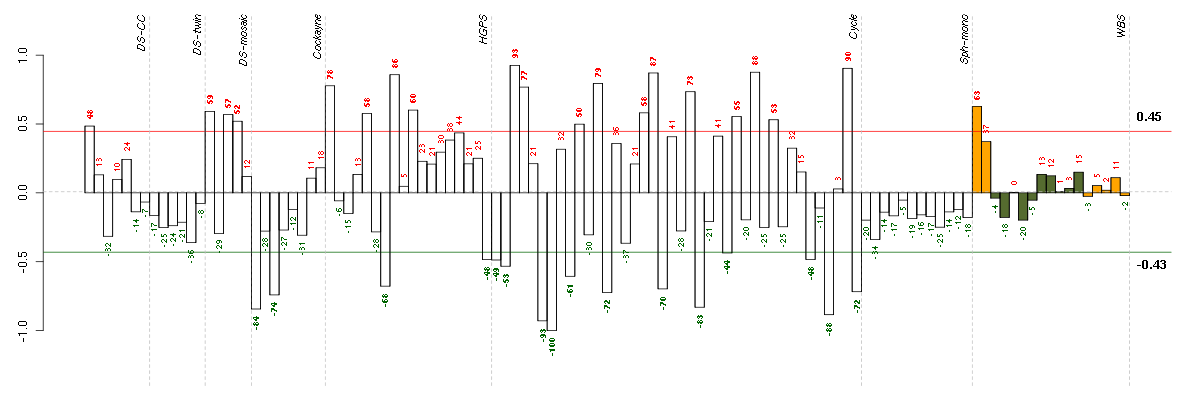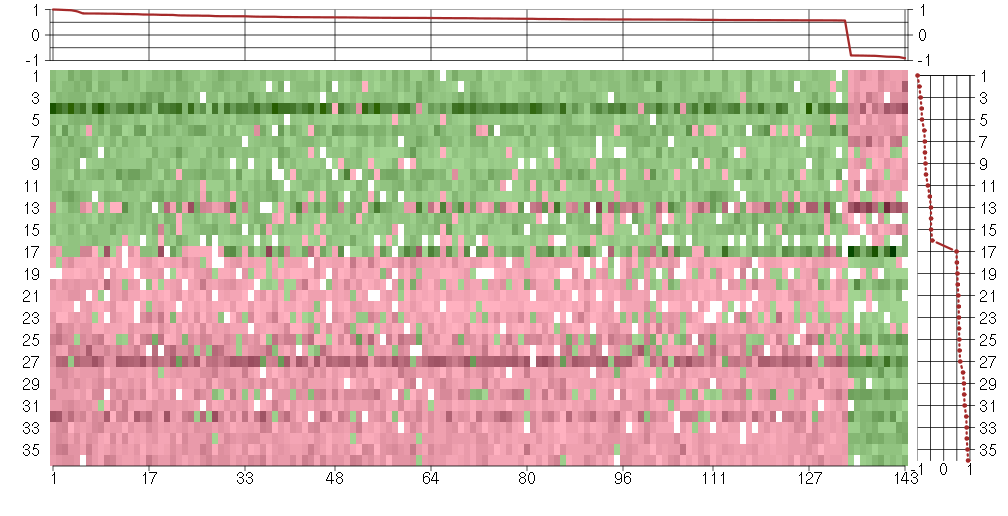

Under-expression is coded with green,
over-expression with red color.



AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: 0.62 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.8 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.6 AESamino-terminal enhancer of split (217729_s_at), score: 0.6 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.73 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.61 AP2A2adaptor-related protein complex 2, alpha 2 subunit (212159_x_at), score: 0.61 APBB1amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) (202652_at), score: 0.66 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.69 ARID1AAT rich interactive domain 1A (SWI-like) (210649_s_at), score: 0.58 ARSAarylsulfatase A (204443_at), score: 0.84 ATN1atrophin 1 (40489_at), score: 0.84 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.72 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.59 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.74 BRD4bromodomain containing 4 (202102_s_at), score: 0.66 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.77 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.68 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.81 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.81 CABIN1calcineurin binding protein 1 (37652_at), score: 0.68 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.67 CALRcalreticulin (200935_at), score: 0.57 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.93 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.82 CENPTcentromere protein T (218148_at), score: 0.58 CHMP5chromatin modifying protein 5 (218085_at), score: -0.85 CHPFchondroitin polymerizing factor (202175_at), score: 0.64 CICcapicua homolog (Drosophila) (212784_at), score: 0.82 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.65 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.8 DBN1drebrin 1 (217025_s_at), score: 0.62 DNM2dynamin 2 (202253_s_at), score: 0.74 DOK4docking protein 4 (209691_s_at), score: 0.62 DOPEY1dopey family member 1 (40612_at), score: -0.86 DUSP7dual specificity phosphatase 7 (214793_at), score: 0.6 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.67 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.61 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.7 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.8 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.61 FOXK2forkhead box K2 (203064_s_at), score: 0.77 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.76 GNAI2guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 (201040_at), score: 0.64 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.83 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.84 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.61 HSF1heat shock transcription factor 1 (202344_at), score: 0.58 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: 0.61 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.73 IGFBP4insulin-like growth factor binding protein 4 (201508_at), score: 0.57 INTS1integrator complex subunit 1 (212212_s_at), score: 0.6 JUNDjun D proto-oncogene (203751_x_at), score: 0.79 JUPjunction plakoglobin (201015_s_at), score: 0.72 KDELR1KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 (200922_at), score: 0.58 KIAA1539KIAA1539 (207765_s_at), score: 0.57 LEPREL2leprecan-like 2 (204854_at), score: 0.7 LMF2lipase maturation factor 2 (212682_s_at), score: 0.66 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.82 LPPR2lipid phosphate phosphatase-related protein type 2 (218509_at), score: 0.69 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: 0.61 MAGOH2mago-nashi homolog 2, proliferation-associated (Drosophila) (217693_x_at), score: -0.81 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.99 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.76 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.65 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.82 MBD3methyl-CpG binding domain protein 3 (41160_at), score: 0.58 MED15mediator complex subunit 15 (222175_s_at), score: 0.58 MGAT4Bmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (220189_s_at), score: 0.67 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.58 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.7 MYO9Bmyosin IXB (217297_s_at), score: 0.61 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.61 NCDNneurochondrin (209556_at), score: 0.63 NCKAP1NCK-associated protein 1 (217465_at), score: -0.91 NCLNnicalin homolog (zebrafish) (222206_s_at), score: 0.58 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: -0.8 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.68 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.61 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: 0.63 PARVBparvin, beta (204629_at), score: 0.79 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.76 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.79 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.59 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.57 PCDHGC3protocadherin gamma subfamily C, 3 (211066_x_at), score: 0.62 PDLIM7PDZ and LIM domain 7 (enigma) (203370_s_at), score: 0.59 PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (207105_s_at), score: 0.67 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 1 PKD1polycystic kidney disease 1 (autosomal dominant) (202328_s_at), score: 0.61 PKN1protein kinase N1 (202161_at), score: 0.67 PLD3phospholipase D family, member 3 (201050_at), score: 0.62 PLSCR3phospholipid scramblase 3 (218828_at), score: 0.63 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.67 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.69 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.72 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.74 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.69 PRKCDprotein kinase C, delta (202545_at), score: 0.64 PTOV1prostate tumor overexpressed 1 (212032_s_at), score: 0.7 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.66 PXNpaxillin (211823_s_at), score: 0.76 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.69 RANBP3RAN binding protein 3 (210120_s_at), score: 0.68 RASA4RAS p21 protein activator 4 (208534_s_at), score: 0.59 RBM15BRNA binding motif protein 15B (202689_at), score: 0.69 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.57 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.83 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: 0.59 SBF1SET binding factor 1 (39835_at), score: 0.66 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.73 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.97 SDAD1SDA1 domain containing 1 (218607_s_at), score: 0.66 SDC3syndecan 3 (202898_at), score: 0.58 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: 0.59 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.61 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.57 SLC4A2solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1) (202111_at), score: 0.59 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.68 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.7 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.84 SORBS3sorbin and SH3 domain containing 3 (209253_at), score: 0.65 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.61 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.84 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.74 SUPT6Hsuppressor of Ty 6 homolog (S. cerevisiae) (208831_x_at), score: 0.64 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.71 TGFB1transforming growth factor, beta 1 (203085_s_at), score: 0.64 TMEM184Ctransmembrane protein 184C (219074_at), score: -0.85 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.63 TSKUtsukushin (218245_at), score: 0.61 TXLNAtaxilin alpha (212300_at), score: 0.59 TYRO3TYRO3 protein tyrosine kinase (211432_s_at), score: 0.58 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.67 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.67 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.98 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.85 XAB2XPA binding protein 2 (218110_at), score: 0.6 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.67 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.72 ZNF282zinc finger protein 282 (212892_at), score: 0.7 ZNF580zinc finger protein 580 (220748_s_at), score: 0.59 ZYXzyxin (200808_s_at), score: 0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |