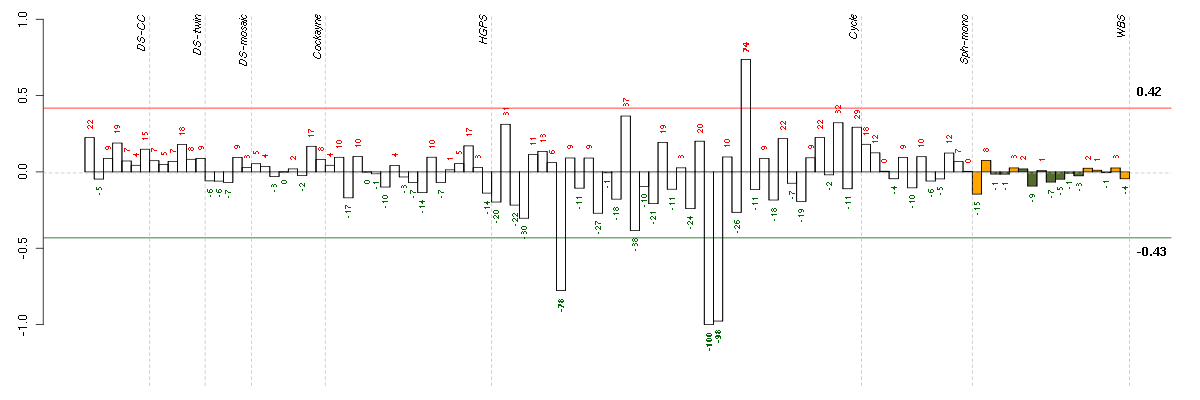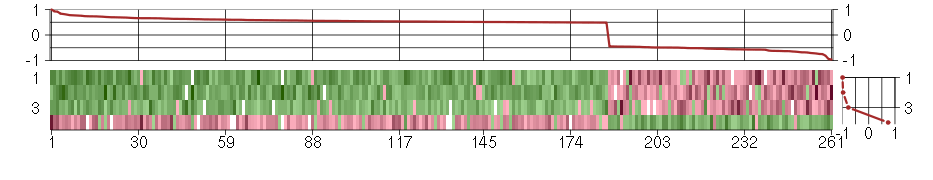

Under-expression is coded with green,
over-expression with red color.


membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or carbohydrate groups.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.

ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.7 ABCC9ATP-binding cassette, sub-family C (CFTR/MRP), member 9 (208561_at), score: -0.56 ACRV1acrosomal vesicle protein 1 (207969_x_at), score: -0.57 ADAMTS20ADAM metallopeptidase with thrombospondin type 1 motif, 20 (220717_at), score: 0.58 ADCY2adenylate cyclase 2 (brain) (217687_at), score: 0.5 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: 0.69 AFTPHaftiphilin (217939_s_at), score: 0.65 AHI1Abelson helper integration site 1 (221569_at), score: -0.49 AKAP6A kinase (PRKA) anchor protein 6 (205359_at), score: -0.52 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: 0.92 AMELXamelogenin (amelogenesis imperfecta 1, X-linked) (208410_x_at), score: 0.55 ANXA10annexin A10 (210143_at), score: -0.52 ARHGEF17Rho guanine nucleotide exchange factor (GEF) 17 (203756_at), score: 0.49 ARHGEF4Rho guanine nucleotide exchange factor (GEF) 4 (205109_s_at), score: 0.52 ASB9ankyrin repeat and SOCS box-containing 9 (205673_s_at), score: -0.65 ASCC2activating signal cointegrator 1 complex subunit 2 (215684_s_at), score: 0.51 ATF6Bactivating transcription factor 6 beta (203168_at), score: 0.49 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: 0.58 ATP2C2ATPase, Ca++ transporting, type 2C, member 2 (214798_at), score: 0.55 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214244_s_at), score: 0.64 B3GALT4UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (210205_at), score: 0.71 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: 0.54 BAGEB melanoma antigen (207712_at), score: -0.46 BEANbrain expressed, associated with Nedd4 (214068_at), score: -0.57 BEND5BEN domain containing 5 (219670_at), score: 0.54 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: -0.48 C17orf80chromosome 17 open reading frame 80 (221048_x_at), score: 0.54 C22orf30chromosome 22 open reading frame 30 (216555_at), score: 0.73 C2orf67chromosome 2 open reading frame 67 (215046_at), score: 0.51 C6orf35chromosome 6 open reading frame 35 (218453_s_at), score: -0.57 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.62 CACNA1Ccalcium channel, voltage-dependent, L type, alpha 1C subunit (211592_s_at), score: 0.76 CACNB1calcium channel, voltage-dependent, beta 1 subunit (206996_x_at), score: 0.52 CADM4cell adhesion molecule 4 (222293_at), score: 0.51 CALML3calmodulin-like 3 (210020_x_at), score: -0.63 CARD8caspase recruitment domain family, member 8 (204950_at), score: 0.51 CASKcalcium/calmodulin-dependent serine protein kinase (MAGUK family) (211208_s_at), score: -0.45 CATSPER2cation channel, sperm associated 2 (217588_at), score: 0.72 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.6 CCNT2cyclin T2 (204645_at), score: -0.49 CD22CD22 molecule (38521_at), score: 0.53 CD320CD320 molecule (218529_at), score: 0.53 CDC42SE1CDC42 small effector 1 (218157_x_at), score: 0.53 CDH18cadherin 18, type 2 (206280_at), score: 0.5 CFHR2complement factor H-related 2 (206910_x_at), score: 0.66 CHKAcholine kinase alpha (204233_s_at), score: 0.56 CHRNA5cholinergic receptor, nicotinic, alpha 5 (206533_at), score: -0.62 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: 0.64 CLCN2chloride channel 2 (213499_at), score: 0.7 COL9A2collagen, type IX, alpha 2 (213622_at), score: 0.66 COPS7BCOP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) (219997_s_at), score: 0.64 CR1complement component (3b/4b) receptor 1 (Knops blood group) (217484_at), score: 0.51 CRISPLD2cysteine-rich secretory protein LCCL domain containing 2 (221541_at), score: 0.63 CTDP1CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 (205035_at), score: 0.52 CTSZcathepsin Z (210042_s_at), score: 0.66 CYBBcytochrome b-245, beta polypeptide (203923_s_at), score: 0.53 CYorf15Bchromosome Y open reading frame 15B (214131_at), score: 0.54 CYP2B6cytochrome P450, family 2, subfamily B, polypeptide 6 (217133_x_at), score: 0.51 CYTH4cytohesin 4 (219183_s_at), score: 0.51 DAPK3death-associated protein kinase 3 (203890_s_at), score: 0.51 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: 0.75 DNAJC2DnaJ (Hsp40) homolog, subfamily C, member 2 (213097_s_at), score: 0.54 DTNAdystrobrevin, alpha (205741_s_at), score: 0.55 EDAectodysplasin A (211130_x_at), score: 0.67 EDNRBendothelin receptor type B (204271_s_at), score: -0.94 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: -0.55 ENPP4ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative function) (204160_s_at), score: -0.49 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: 0.52 EPHB6EPH receptor B6 (204718_at), score: 0.53 EPN1epsin 1 (221141_x_at), score: 0.68 EPOerythropoietin (217254_s_at), score: 1 EPS8L1EPS8-like 1 (218778_x_at), score: 0.6 ETNK2ethanolamine kinase 2 (219268_at), score: 0.52 EXOSC2exosome component 2 (209527_at), score: -0.49 FBRSfibrosin (218255_s_at), score: 0.51 FBXW7F-box and WD repeat domain containing 7 (218751_s_at), score: 0.49 FEM1Cfem-1 homolog c (C. elegans) (213341_at), score: -0.54 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.63 FLCNfolliculin (215645_at), score: 0.63 FRAT2frequently rearranged in advanced T-cell lymphomas 2 (209864_at), score: -0.63 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: 0.54 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.5 G6PC2glucose-6-phosphatase, catalytic, 2 (221453_at), score: 0.55 GABRG3gamma-aminobutyric acid (GABA) A receptor, gamma 3 (216895_at), score: -0.75 GASTgastrin (208138_at), score: 0.56 GCHFRGTP cyclohydrolase I feedback regulator (204867_at), score: 0.74 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: 0.51 GNAO1guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O (204762_s_at), score: -0.57 GPM6Bglycoprotein M6B (209170_s_at), score: -0.47 GPR144G protein-coupled receptor 144 (216289_at), score: 0.59 GPR85G protein-coupled receptor 85 (219898_at), score: 0.58 GRM1glutamate receptor, metabotropic 1 (210939_s_at), score: 0.51 GRWD1glutamate-rich WD repeat containing 1 (221549_at), score: 0.61 GTF2H4general transcription factor IIH, polypeptide 4, 52kDa (203577_at), score: 0.58 GYG2glycogenin 2 (215695_s_at), score: 0.49 HAB1B1 for mucin (215778_x_at), score: 0.53 HCRP1hepatocellular carcinoma-related HCRP1 (216176_at), score: 0.6 HDAC5histone deacetylase 5 (202455_at), score: 0.69 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.5 HHIPL2HHIP-like 2 (220283_at), score: 0.6 HINFPhistone H4 transcription factor (206495_s_at), score: 0.66 HIST1H2ALhistone cluster 1, H2al (214554_at), score: -0.96 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.57 HPNhepsin (204934_s_at), score: 0.61 IL22RA1interleukin 22 receptor, alpha 1 (220056_at), score: 0.69 IL33interleukin 33 (209821_at), score: -0.57 IL7interleukin 7 (206693_at), score: 0.53 INHAinhibin, alpha (210141_s_at), score: 0.51 IRS4insulin receptor substrate 4 (207403_at), score: 0.55 ISOC2isochorismatase domain containing 2 (218893_at), score: 0.59 ITGB3integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) (204628_s_at), score: 0.55 ITGB8integrin, beta 8 (211488_s_at), score: -0.69 JARID1Cjumonji, AT rich interactive domain 1C (202383_at), score: 0.55 KCNQ3potassium voltage-gated channel, KQT-like subfamily, member 3 (206573_at), score: -0.74 KIAA0415KIAA0415 (209912_s_at), score: 0.55 KIAA1324KIAA1324 (221874_at), score: -0.54 KIAA1659KIAA1659 protein (215674_at), score: 0.49 KIF13Bkinesin family member 13B (202962_at), score: 0.5 KIF26Bkinesin family member 26B (220002_at), score: 0.6 KLHL5kelch-like 5 (Drosophila) (220682_s_at), score: 0.61 KLK13kallikrein-related peptidase 13 (205783_at), score: -0.57 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (205307_s_at), score: -0.46 KRT86keratin 86 (215189_at), score: 0.84 KSR1kinase suppressor of ras 1 (213769_at), score: 0.51 KTELC1KTEL (Lys-Tyr-Glu-Leu) containing 1 (218587_s_at), score: 0.61 LAMB4laminin, beta 4 (215516_at), score: -0.54 LOC100129624hypothetical LOC100129624 (210748_at), score: 0.5 LOC100133572similar to seven transmembrane helix receptor (215770_at), score: -0.56 LOC100133748similar to GTF2IRD2 protein (215569_at), score: 0.53 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.66 LOC23117PI-3-kinase-related kinase SMG-1 isoform 1 homolog (211996_s_at), score: 0.55 LOC91316glucuronidase, beta/ immunoglobulin lambda-like polypeptide 1 pseudogene (215816_at), score: 0.56 LRP3low density lipoprotein receptor-related protein 3 (204381_at), score: 0.59 LRRN2leucine rich repeat neuronal 2 (216164_at), score: 0.66 LSRlipolysis stimulated lipoprotein receptor (208190_s_at), score: -0.62 LUZP1leucine zipper protein 1 (221832_s_at), score: 0.54 LY75lymphocyte antigen 75 (205668_at), score: -0.51 MACROD1MACRO domain containing 1 (219188_s_at), score: 0.62 MAPK12mitogen-activated protein kinase 12 (206106_at), score: 0.5 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.54 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: -0.56 MCF2L2MCF.2 cell line derived transforming sequence-like 2 (215112_x_at), score: -0.5 MED31mediator complex subunit 31 (219318_x_at), score: -0.46 MGAT4Cmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) (207447_s_at), score: -0.73 MGC13053hypothetical MGC13053 (211775_x_at), score: -0.54 MIA2melanoma inhibitory activity 2 (221177_at), score: -0.56 MID2midline 2 (208384_s_at), score: 0.52 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.45 MOCS3molybdenum cofactor synthesis 3 (206141_at), score: 0.55 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: -0.68 MTHFSDmethenyltetrahydrofolate synthetase domain containing (218879_s_at), score: 0.66 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.55 NAPGN-ethylmaleimide-sensitive factor attachment protein, gamma (210048_at), score: 0.58 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.7 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.61 NPEPL1aminopeptidase-like 1 (89476_r_at), score: 0.5 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.57 NUAK1NUAK family, SNF1-like kinase, 1 (204589_at), score: 0.53 NYXnyctalopin (221684_s_at), score: -0.68 OR1E2olfactory receptor, family 1, subfamily E, member 2 (208587_s_at), score: 0.53 OR3A2olfactory receptor, family 3, subfamily A, member 2 (221386_at), score: 0.62 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: 0.58 OVOL2ovo-like 2 (Drosophila) (211778_s_at), score: 0.63 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: 0.52 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: 0.5 PAX4paired box 4 (207867_at), score: -0.49 PCSK1proprotein convertase subtilisin/kexin type 1 (205825_at), score: -0.56 PDE1Cphosphodiesterase 1C, calmodulin-dependent 70kDa (207303_at), score: 0.57 PDK3pyruvate dehydrogenase kinase, isozyme 3 (221957_at), score: -0.67 PEX16peroxisomal biogenesis factor 16 (221604_s_at), score: 0.51 PEX7peroxisomal biogenesis factor 7 (205420_at), score: -0.45 PF4V1platelet factor 4 variant 1 (207815_at), score: -0.52 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.48 PKNOX1PBX/knotted 1 homeobox 1 (221883_at), score: 0.6 PLA2G7phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) (206214_at), score: 0.57 PMF1polyamine-modulated factor 1 (202337_at), score: 0.53 PPP1R14Dprotein phosphatase 1, regulatory (inhibitor) subunit 14D (220082_at), score: 0.56 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.57 PROX1prospero homeobox 1 (207401_at), score: -0.45 PTP4A3protein tyrosine phosphatase type IVA, member 3 (209695_at), score: 0.57 PTPRRprotein tyrosine phosphatase, receptor type, R (210675_s_at), score: 0.5 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.58 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -0.51 RBAKRB-associated KRAB zinc finger (219214_s_at), score: 0.51 RHBDF2rhomboid 5 homolog 2 (Drosophila) (219202_at), score: -0.46 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: 0.5 RNF121ring finger protein 121 (219021_at), score: 0.57 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: 0.51 RPAP2RNA polymerase II associated protein 2 (219504_s_at), score: 0.61 RPL14ribosomal protein L14 (219138_at), score: 0.53 RPL27Aribosomal protein L27a (212044_s_at), score: 0.59 SAA4serum amyloid A4, constitutive (207096_at), score: 0.5 SAMD14sterile alpha motif domain containing 14 (213866_at), score: 0.73 SARM1sterile alpha and TIR motif containing 1 (213259_s_at), score: 0.8 SCN10Asodium channel, voltage-gated, type X, alpha subunit (208578_at), score: -0.54 SECISBP2SECIS binding protein 2 (218265_at), score: 0.5 SEMA4Gsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G (219194_at), score: -0.45 SEPT5septin 5 (209767_s_at), score: 0.55 SETD1ASET domain containing 1A (213202_at), score: 0.5 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.51 SHPKsedoheptulokinase (219713_at), score: 0.62 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: 0.66 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: -0.63 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.57 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: -0.53 SLC35D2solute carrier family 35, member D2 (213082_s_at), score: -0.5 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.77 SMA4glucuronidase, beta pseudogene (215599_at), score: -0.65 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: 0.52 SP140SP140 nuclear body protein (207777_s_at), score: -0.52 SPAG1sperm associated antigen 1 (210117_at), score: -0.49 SPAG8sperm associated antigen 8 (206816_s_at), score: 0.56 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.93 SUV39H2suppressor of variegation 3-9 homolog 2 (Drosophila) (219262_at), score: -0.47 SV2Asynaptic vesicle glycoprotein 2A (203069_at), score: 0.5 SYT1synaptotagmin I (203999_at), score: -0.81 SYT5synaptotagmin V (206161_s_at), score: 0.63 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.63 TBL3transducin (beta)-like 3 (209820_s_at), score: 0.52 TBX10T-box 10 (207689_at), score: 0.77 TCF20transcription factor 20 (AR1) (212931_at), score: 0.73 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: 0.52 TICAM1toll-like receptor adaptor molecule 1 (213191_at), score: 0.53 TMEM184Ctransmembrane protein 184C (219074_at), score: 0.49 TMEM62transmembrane protein 62 (218776_s_at), score: -0.58 TMEM63Atransmembrane protein 63A (202700_s_at), score: 0.54 TMOD2tropomodulin 2 (neuronal) (219701_at), score: -0.49 TMPRSS6transmembrane protease, serine 6 (214955_at), score: 0.78 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.49 TNFRSF8tumor necrosis factor receptor superfamily, member 8 (206729_at), score: 0.82 TP53TG1TP53 target 1 (non-protein coding) (210241_s_at), score: 0.5 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.63 TRAF4TNF receptor-associated factor 4 (202871_at), score: 0.61 TRIM2tripartite motif-containing 2 (202341_s_at), score: 0.51 TRIM33tripartite motif-containing 33 (210266_s_at), score: 0.49 TRPM3transient receptor potential cation channel, subfamily M, member 3 (220463_at), score: 0.51 TSPAN9tetraspanin 9 (205665_at), score: 0.6 TXNL4Bthioredoxin-like 4B (218794_s_at), score: 0.54 TYMPthymidine phosphorylase (217497_at), score: 0.52 UGT2A1UDP glucuronosyltransferase 2 family, polypeptide A1 (207958_at), score: 0.73 UNGuracil-DNA glycosylase (202330_s_at), score: -0.46 USP20ubiquitin specific peptidase 20 (203965_at), score: 0.5 UTF1undifferentiated embryonic cell transcription factor 1 (208275_x_at), score: 0.62 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.53 WDR4WD repeat domain 4 (221632_s_at), score: 0.49 WHAMML1WAS protein homolog associated with actin, golgi membranes and microtubules-like 1 (213908_at), score: -0.49 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.72 ZBTB10zinc finger and BTB domain containing 10 (219312_s_at), score: -0.54 ZCCHC14zinc finger, CCHC domain containing 14 (212655_at), score: 0.49 ZGPATzinc finger, CCCH-type with G patch domain (221848_at), score: 0.67 ZNF193zinc finger protein 193 (205181_at), score: 0.54 ZNF434zinc finger protein 434 (218937_at), score: 0.53 ZNF467zinc finger protein 467 (214746_s_at), score: -0.71 ZNF516zinc finger protein 516 (203604_at), score: 0.49 ZNF643zinc finger protein 643 (207219_at), score: 0.49 ZNF671zinc finger protein 671 (219849_at), score: 0.49 ZNF749zinc finger protein 749 (215289_at), score: -0.49 ZNF768zinc finger protein 768 (218916_at), score: 0.65 ZNF787zinc finger protein 787 (213402_at), score: 0.5 ZNF814zinc finger protein 814 (60794_f_at), score: -0.71 ZNF839zinc finger protein 839 (221709_s_at), score: 0.58
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |