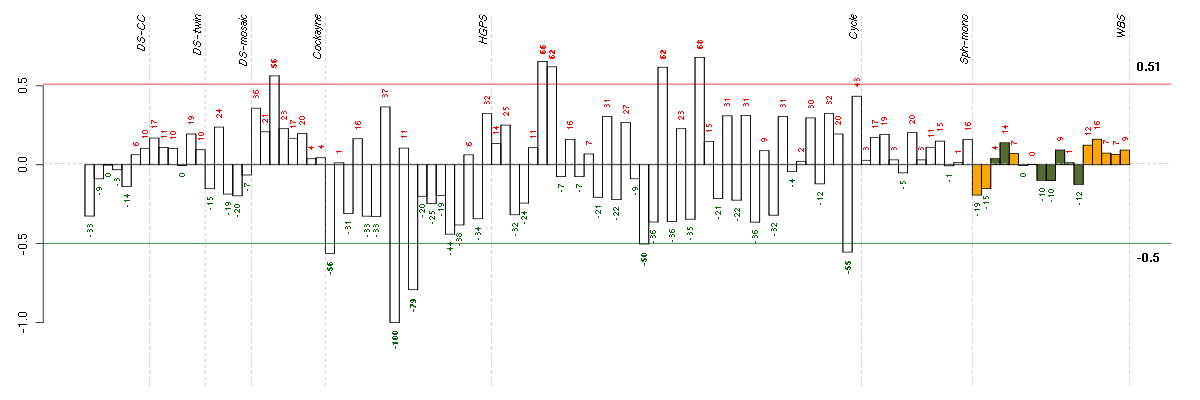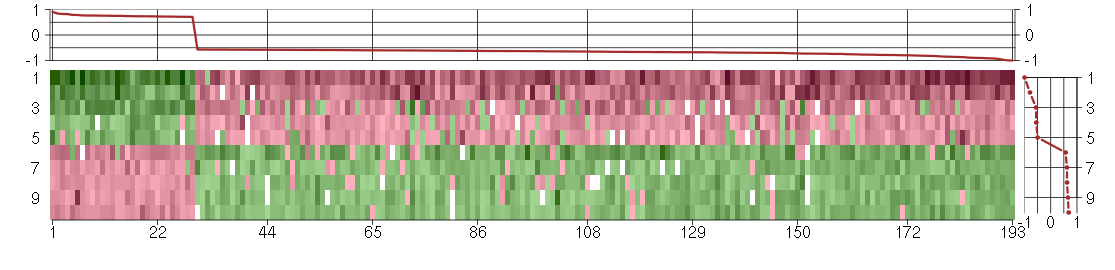

Under-expression is coded with green,
over-expression with red color.



ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: -0.58 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.7 AESamino-terminal enhancer of split (217729_s_at), score: -0.63 AK3L1adenylate kinase 3-like 1 (204348_s_at), score: -0.57 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: -0.66 ANAPC10anaphase promoting complex subunit 10 (207845_s_at), score: 0.72 AP2A2adaptor-related protein complex 2, alpha 2 subunit (212159_x_at), score: -0.58 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: -0.72 ARID1AAT rich interactive domain 1A (SWI-like) (210649_s_at), score: -0.6 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: 0.76 ARSAarylsulfatase A (204443_at), score: -0.58 ASF1AASF1 anti-silencing function 1 homolog A (S. cerevisiae) (203428_s_at), score: 0.75 ATF2activating transcription factor 2 (212984_at), score: 0.73 ATN1atrophin 1 (40489_at), score: -0.76 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: -0.83 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: -0.8 BIN1bridging integrator 1 (210201_x_at), score: -0.59 BLCAPbladder cancer associated protein (201032_at), score: -0.59 BRD9bromodomain containing 9 (220155_s_at), score: -0.67 BRMS1breast cancer metastasis suppressor 1 (215631_s_at), score: -0.67 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.67 C14orf104chromosome 14 open reading frame 104 (219166_at), score: 0.75 C19orf22chromosome 19 open reading frame 22 (221764_at), score: -0.61 C22orf29chromosome 22 open reading frame 29 (219560_at), score: -0.59 CABIN1calcineurin binding protein 1 (37652_at), score: -0.58 CAMK2N1calcium/calmodulin-dependent protein kinase II inhibitor 1 (218309_at), score: -0.63 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: -0.63 CCNE1cyclin E1 (213523_at), score: -0.64 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.78 CDKN1Bcyclin-dependent kinase inhibitor 1B (p27, Kip1) (209112_at), score: 0.91 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: -0.82 CDT1chromatin licensing and DNA replication factor 1 (209832_s_at), score: -0.67 CENPTcentromere protein T (218148_at), score: -0.77 CHMP1Achromatin modifying protein 1A (201933_at), score: -0.59 CHPFchondroitin polymerizing factor (202175_at), score: -0.64 CHST3carbohydrate (chondroitin 6) sulfotransferase 3 (209834_at), score: -0.78 CICcapicua homolog (Drosophila) (212784_at), score: -0.63 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: -0.62 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: -0.59 CLUclusterin (208791_at), score: -0.65 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.67 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: -0.79 COL8A2collagen, type VIII, alpha 2 (221900_at), score: -0.68 COMPcartilage oligomeric matrix protein (205713_s_at), score: -0.65 COPS7ACOP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) (209029_at), score: -0.67 CTDSPLCTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like (201904_s_at), score: -0.58 DCTN1dynactin 1 (p150, glued homolog, Drosophila) (201082_s_at), score: -0.67 DGCR8DiGeorge syndrome critical region gene 8 (218650_at), score: -0.58 DNM1dynamin 1 (215116_s_at), score: -0.61 DNM2dynamin 2 (202253_s_at), score: -0.61 DOK4docking protein 4 (209691_s_at), score: -0.7 EPS15epidermal growth factor receptor pathway substrate 15 (217887_s_at), score: 0.77 F2Rcoagulation factor II (thrombin) receptor (203989_x_at), score: 0.75 FAM134Cfamily with sequence similarity 134, member C (212697_at), score: -0.57 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: -0.77 FAM38Afamily with sequence similarity 38, member A (202771_at), score: -0.59 FASNfatty acid synthase (212218_s_at), score: -0.84 FBXO17F-box protein 17 (220233_at), score: -0.61 FBXO3F-box protein 3 (218432_at), score: 0.72 FHL1four and a half LIM domains 1 (201539_s_at), score: 0.71 FHOD3formin homology 2 domain containing 3 (218980_at), score: -0.61 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: -0.62 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.73 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: 0.74 FOXK2forkhead box K2 (203064_s_at), score: -0.68 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: -0.91 FXYD5FXYD domain containing ion transport regulator 5 (218084_x_at), score: -0.57 GAAglucosidase, alpha; acid (202812_at), score: -0.7 GAS7growth arrest-specific 7 (202191_s_at), score: -0.63 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: -0.87 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: -0.73 GPR153G protein-coupled receptor 153 (221902_at), score: -0.57 GPR172AG protein-coupled receptor 172A (222155_s_at), score: -0.6 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: -0.74 GYS1glycogen synthase 1 (muscle) (201673_s_at), score: -0.65 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.58 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.79 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: -0.64 HOXC11homeobox C11 (206745_at), score: -0.74 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.62 IFT57intraflagellar transport 57 homolog (Chlamydomonas) (218100_s_at), score: 0.73 IGFBP4insulin-like growth factor binding protein 4 (201508_at), score: -0.64 IL11interleukin 11 (206924_at), score: -0.59 IL17RAinterleukin 17 receptor A (205707_at), score: -0.68 IL4Rinterleukin 4 receptor (203233_at), score: -0.57 IQSEC1IQ motif and Sec7 domain 1 (203907_s_at), score: -0.68 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: -0.69 ITGA5integrin, alpha 5 (fibronectin receptor, alpha polypeptide) (201389_at), score: -0.6 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: -0.7 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: -0.66 LMF2lipase maturation factor 2 (212682_s_at), score: -0.69 LOC284440hypothetical LOC284440 (216126_at), score: 0.71 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.61 LOC90379hypothetical protein BC002926 (221849_s_at), score: -0.63 LPPR2lipid phosphate phosphatase-related protein type 2 (218509_at), score: -0.62 LRFN4leucine rich repeat and fibronectin type III domain containing 4 (219491_at), score: -0.79 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200784_s_at), score: -0.58 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.85 MAP3K3mitogen-activated protein kinase kinase kinase 3 (203514_at), score: -0.58 MAP7D1MAP7 domain containing 1 (217943_s_at), score: -0.9 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: -0.86 MED15mediator complex subunit 15 (222175_s_at), score: -0.6 MED4mediator complex subunit 4 (217843_s_at), score: 0.72 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: -0.62 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: -0.86 MTA2metastasis associated 1 family, member 2 (203442_x_at), score: -0.6 MYO9Bmyosin IXB (217297_s_at), score: -0.62 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.93 NCKAP1NCK-associated protein 1 (217465_at), score: 0.79 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: -0.68 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: 0.76 NET1neuroepithelial cell transforming 1 (201830_s_at), score: -0.63 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.76 NFYBnuclear transcription factor Y, beta (218129_s_at), score: 0.71 NKX3-1NK3 homeobox 1 (209706_at), score: -0.88 NR1D1nuclear receptor subfamily 1, group D, member 1 (31637_s_at), score: -0.69 OLFM1olfactomedin 1 (213131_at), score: -0.73 ORAI2ORAI calcium release-activated calcium modulator 2 (218812_s_at), score: -0.58 OTUD3OTU domain containing 3 (213216_at), score: -0.63 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: -0.64 PARVBparvin, beta (204629_at), score: -0.68 PCDHG@protocadherin gamma cluster (215836_s_at), score: -0.78 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.97 PCDHGC3protocadherin gamma subfamily C, 3 (211066_x_at), score: -0.89 PEX26peroxisomal biogenesis factor 26 (219180_s_at), score: -0.7 PHF20PHD finger protein 20 (209422_at), score: 0.73 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.99 PKD1polycystic kidney disease 1 (autosomal dominant) (202328_s_at), score: -0.58 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: -0.73 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: -0.74 PLEKHJ1pleckstrin homology domain containing, family J member 1 (218290_at), score: -0.61 PLSCR3phospholipid scramblase 3 (218828_at), score: -0.58 PLXNA3plexin A3 (203623_at), score: -0.64 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: -0.59 POLIpolymerase (DNA directed) iota (219317_at), score: 0.76 POLSpolymerase (DNA directed) sigma (202466_at), score: -0.59 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.81 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: -0.6 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: -0.81 PRLRprolactin receptor (216638_s_at), score: -0.58 PSD4pleckstrin and Sec7 domain containing 4 (203317_at), score: -0.64 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.63 PTK7PTK7 protein tyrosine kinase 7 (207011_s_at), score: -0.58 RAB11FIP5RAB11 family interacting protein 5 (class I) (210879_s_at), score: -0.6 RAB15RAB15, member RAS onocogene family (221810_at), score: -0.73 RAD17RAD17 homolog (S. pombe) (207405_s_at), score: 0.73 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.67 RASA4RAS p21 protein activator 4 (208534_s_at), score: -0.67 RBM15BRNA binding motif protein 15B (202689_at), score: -0.63 RNF126ring finger protein 126 (205748_s_at), score: -0.66 RSL24D1ribosomal L24 domain containing 1 (217915_s_at), score: 0.82 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.9 SDC4syndecan 4 (202071_at), score: -0.7 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: -0.58 SETBP1SET binding protein 1 (205933_at), score: -0.7 SF1splicing factor 1 (208313_s_at), score: -0.67 SFRP1secreted frizzled-related protein 1 (202036_s_at), score: -0.57 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: -0.62 SIPA1L3signal-induced proliferation-associated 1 like 3 (213600_at), score: -0.76 SLC25A22solute carrier family 25 (mitochondrial carrier: glutamate), member 22 (218725_at), score: -0.65 SLC6A10Psolute carrier family 6 (neurotransmitter transporter, creatine), member 10 (pseudogene) (215812_s_at), score: -0.6 SLC6A8solute carrier family 6 (neurotransmitter transporter, creatine), member 8 (202219_at), score: -0.6 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: -0.73 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: -0.67 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.72 SPHK1sphingosine kinase 1 (219257_s_at), score: -0.65 SPON1spondin 1, extracellular matrix protein (209436_at), score: -0.61 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: -0.69 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: -0.69 STC1stanniocalcin 1 (204597_x_at), score: -0.6 STK10serine/threonine kinase 10 (203047_at), score: -0.57 STRN4striatin, calmodulin binding protein 4 (217903_at), score: -0.82 STXBP3syntaxin binding protein 3 (203310_at), score: 0.72 SUPT6Hsuppressor of Ty 6 homolog (S. cerevisiae) (208831_x_at), score: -0.69 SUSD5sushi domain containing 5 (214954_at), score: 0.82 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.8 TBC1D10BTBC1 domain family, member 10B (220947_s_at), score: -0.65 TGFAtransforming growth factor, alpha (205016_at), score: -0.78 TGFB1transforming growth factor, beta 1 (203085_s_at), score: -0.61 TMEM123transmembrane protein 123 (211967_at), score: 0.84 TMEM158transmembrane protein 158 (213338_at), score: -0.77 TNK2tyrosine kinase, non-receptor, 2 (203839_s_at), score: -0.66 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.73 TRMT11tRNA methyltransferase 11 homolog (S. cerevisiae) (218877_s_at), score: 0.76 TSKUtsukushin (218245_at), score: -0.64 TXLNAtaxilin alpha (212300_at), score: -0.92 UBE2Oubiquitin-conjugating enzyme E2O (218141_at), score: -0.66 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: -0.63 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.71 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.67 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: 0.74 ZNF277zinc finger protein 277 (218645_at), score: 0.76 ZNF536zinc finger protein 536 (206403_at), score: -1
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |